Predicting whether a clinical trial will succeed or fail is notoriously difficult. Even experienced pharmaceutical analysts struggle with accuracy rates much better than a coin flip. But what if we could train an AI model to learn from thousands of past trials and improve its predictions?
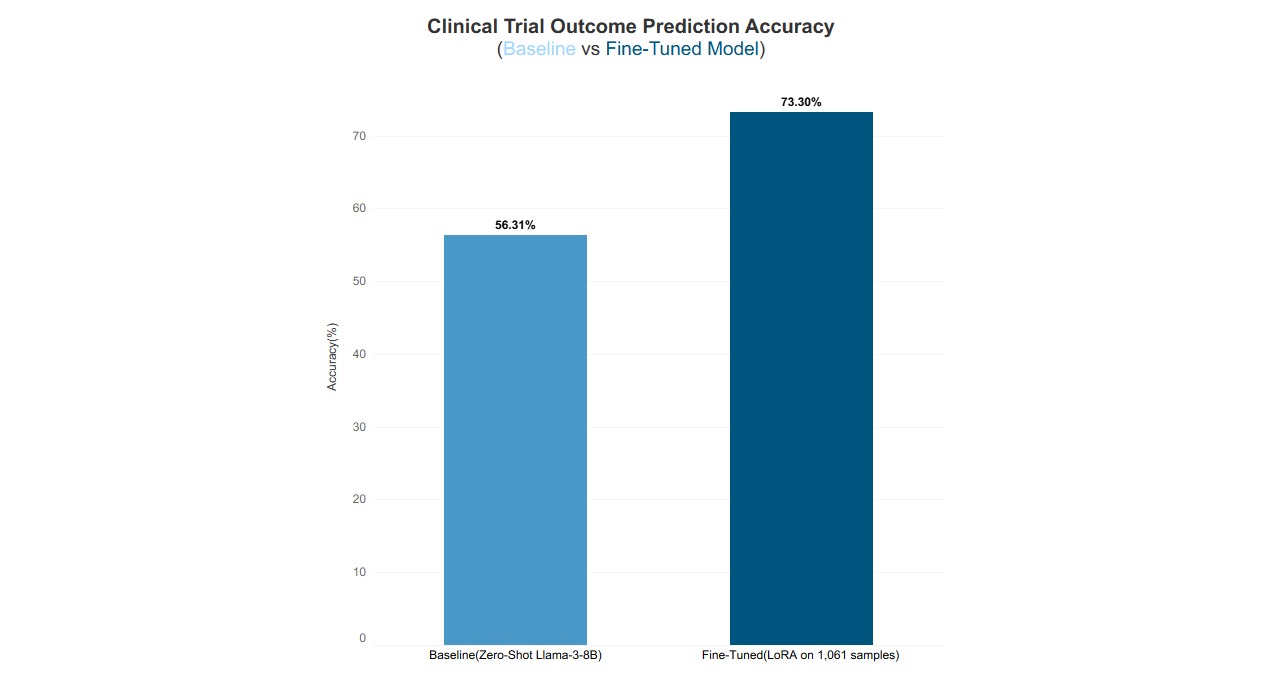
I recently built a dataset of 1,366 clinical trial predictions and fine-tuned an 8B parameter language model to predict trial outcomes. This resulted to in a jump from 56% accuracy (barely better than guessing) to 73% accuracy, a 30% relative improvement. Here’s how I did it, what I learned, and why this matters for anyone working with prediction tasks.
The pharmaceutical industry runs on uncertainty. When Eli Lilly announces a Phase 3 trial for a new obesity drug, analysts, investors, and competitors all ask the same question: Will it succeed?
Traditionally, answering this question requires:
Deep domain expertise in pharmacology
Knowledge of the company’s track record
Understanding of regulatory pathways
Access to clinical trial databases
Lots of time to research each case
Even then, human experts achieve modest accuracy. The question I wanted to answer is, could an AI model learn these patterns automatically from historical data?
The biggest hurdle in building prediction models is getting labeled training data. Hiring medical experts to label thousands of clinical trial outcomes would cost tens of thousands of dollars and take months.
This is where I discovered Lightning Rod’s approach to data generation. Instead of manual labelling, their SDK uses what they call the “Future-as-Label” methodology. This approach makes the future outcome of a historical event its label.
Here’s how it works:
Find old news: Articles from 2023 about clinical trials starting
Generate questions: “Will Novo Nordisk’s Phase 3 trial meet endpoints by Q4 2024?”
Auto-label outcomes: Search recent news (late 2024/2025) to find what actually happened
Build dataset: Pair questions with verified outcomes
It does this without the need for human labellers. The Lightning Rod Python SDK automatically finds the answers by searching for what happened later.
Using Lightning Rod’s Python SDK, I generated the dataset with a simple pipeline:
from lightningrod import QuestionPipeline, NewsSeedGenerator, WebSearchLabeler pipeline = QuestionPipeline( seed_generator=NewsSeedGenerator( start_date=datetime(2023, 1, 1), end_date=datetime(2024, 12, 31), search_query=["clinical trial Phase 3", "FDA approval"] ), question_generator=ForwardLookingQuestionGenerator( instructions="Generate binary questions about trial outcomes", examples=[ "Will Eli Lilly's obesity drug trial meet endpoints by Q4 2024?", "Will the FDA approve Drug X by June 2024?" ] ), labeler=WebSearchLabeler(confidence_threshold=0.7) ) dataset = lr.transforms.run(pipeline, max_questions=2000)
The SDK pulled news articles about clinical trials, generated forward-looking questions, and then searched for later outcomes. In about 10 minutes of compute time, I had 1,882 questions, with 72.6% successfully labeled, giving me 1,366 high-quality training examples.

Each example looked like this:
Question: "Will Novo Nordisk's CagriSema Phase 3 trial meet its primary endpoints by December 31, 2024?" Answer: YES (1) Confidence: 0.98
The labels weren’t guesses; they were verified facts from published trial results and FDA announcements.
I split the data into training (85%) and test (15%) sets, then ran two experiments:
First, I tested Llama-3–8B without any training. I gave each question and asked it to predict 0 (failure) or 1 (success).
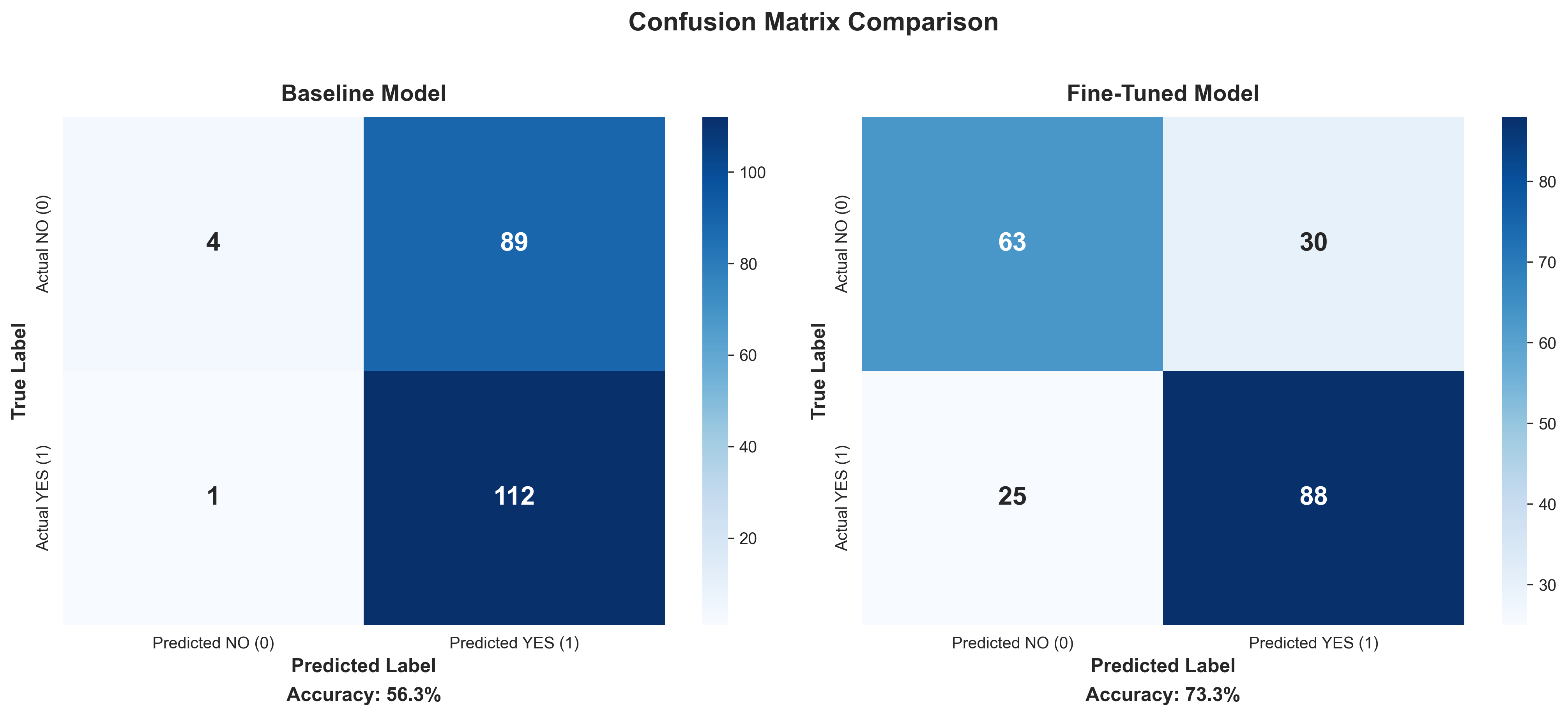
Result: 56.3% Accuracy
The model was essentially guessing, with a slight optimistic bias (it predicted “success” too often). I wasn’t surprised because the base model has no special knowledge of pharmaceutical industry patterns.
Next, I fine-tuned the model using LoRA (Low-Rank Adaptation) on the training data. LoRA is a parameter-efficient method that adds small adapter layers instead of retraining the entire model.
The setup I used:
Model: Llama-3–8B with 4-bit quantization
Method: LoRA fine-tuning via the Unsloth library
Hardware: Free Google Colab T4 GPU
Training time: ~21 minutes (3 epochs)
Trainable parameters: Only 16M (0.2% of the model)
Result: 73.3% Accuracy
The fine-tuned model correctly answered 151 out of 206 test questions, achieving 73.3% accuracy. This represents a 17-percentage-point improvement over the baseline, a 30% relative performance gain achieved in just 21 minutes of training. Notably, this was done using only 0.2% of the model’s parameters over 3 training epochs, demonstrating highly efficient improvement with minimal compute.
The most interesting part wasn’t just the numbers; it was understanding what patterns the model discovered in the data.
The model learned that pharmaceutical companies have different success rates. Questions mentioning Eli Lilly, Novo Nordisk, or Merck were more likely to be “YES” (success), while smaller biotech startups showed higher failure rates.
This makes sense because established companies have more resources, experience, and proven track records. The model picked this up automatically from the data.
Obesity and diabetes drugs showed ~68% success rates in the training data, while oncology trials succeeded only ~48% of the time. The model learned these differences without being explicitly told.
Cancer is harder to treat. Metabolic diseases have clearer biomarkers. The model internalized these domain patterns solely from examples.
One surprising discovery: the model learned to spot unrealistic timelines.
Example questions the model corrected:
“Will [Small Biotech] complete Phase 3 in 6 months?” → Predicted NO (correctly)
“Will [Unproven Drug] get FDA approval in 3 months?” → Predicted NO (correctly)
The baseline model didn’t know that Phase 3 trials typically take 18–24 months. The fine-tuned version learned this pattern from the data.
The baseline model showed an optimistic bias, predicting “success” 63% of the time. The fine-tuned model was better calibrated at 52%, closer to the actual distribution.
More importantly, it learned to identify red flags like:
Aggressive timelines
Unproven mechanisms
Companies with poor track records
Challenging therapeutic areas
All five examples of “most improved” predictions involved the baseline incorrectly predicting success, while the fine-tuned model correctly predicted failure.
While this experiment focused on pharmaceutical trial outcomes, the real contribution goes beyond healthcare. It demonstrates a practical, repeatable workflow for building specialized prediction models from real-world data.
At its core, the approach is straightforward:
This same approach could work for:
Any domain with historical news or announcements, clear and verifiable outcomes, and sufficient examples can benefit from Lightning Rod’s Future-as-Label methodology.
It’s not limited to clinical trials; it serves as a scalable template for temporal prediction tasks across industries.
For developers interested in reproducing this:
The full code and dataset are available on GitHub, and the dataset is published on Hugging Face for anyone who wants to reproduce or build on this work.
This isn’t a perfect crystal ball. The model still struggles with:
The 73% accuracy is a meaningful improvement over guessing, but it’s not prophecy. Think of it as moving from “coin flip” to “informed probability estimate.” For context, even experienced pharmaceutical analysts struggle to achieve accuracy above 65–70% in predicting trial outcomes.
Future improvements could include:
Three lessons from this project:
1. Automated labeling scales: Manually labeling 1,366 examples would have taken weeks and cost thousands. Lightning Rod’s Future-as-Label approach did it in 3 minutes.
2. Small models can specialize:* You don’t need GPT-4 or Claude for domain-specific tasks. An 8B model, fine-tuned on focused data, achieved 73% accuracy on a challenging prediction problem.
3. Historical data contains learnable patterns: Company track records, therapeutic area success rates, and timeline realism all emerged naturally from the training data. The model discovered what experts know from experience.
The tools I used are all publicly available:
Lightning Rod SDK: Open-source Python library for dataset generation
Pre-trained Model: Skip training, use the fine-tuned model directly via Hugging Face
Unsloth: Free library for efficient LoRA fine-tuning
Google Colab: Free GPU access for training
Hugging Face: Free dataset and model hosting
The barrier to entry for specialized AI models has never been lower. If you have a prediction task with historical data, you can build a custom model in a weekend.
The future of AI isn’t just giant general-purpose models; it’s also specialized models trained on focused, high-quality datasets for specific domains. This experiment is one example of what becomes possible when you combine automated data generation with efficient fine-tuning.
What prediction task would you build a model for?
The dataset is available on Hugging Face, and all code is on GitHub. Special thanks to Lightning Rod Labs for their SDK that made this project possible.
Resources:
Interested in building prediction datasets? Check out Lightning Rod or explore their examples on Hugging Face