Abstract
This project introduces a Retrieval-Augmented Generation (RAG) system designed to support cancer research by enabling interactive exploration of text-based literature. Built with ChromaDB for semantic search and Streamlit for an intuitive chat interface, the system allows researchers to load plain text files containing cancer-related studies and ask natural language questions. By combining document retrieval with modern large language models (LLMs) via Groq’s API, the assistant generates contextualized answers grounded in the uploaded research. This project demonstrates how lightweight RAG systems can improve access to biomedical knowledge and serve as an educational example for researchers, students, and AI practitioners.
Introduction
Artificial Intelligence (AI) has rapidly transformed the way researchers interact with scientific literature. In cancer research, where new findings emerge daily, the challenge often lies not in accessing data but in extracting meaningful insights from vast amounts of text. Traditional keyword searches lack the ability to capture context and nuance, leaving researchers with incomplete or irrelevant results.
This project presents a practical solution: a Retrieval-Augmented Generation (RAG) assistant that allows users to upload cancer-related documents and query them interactively. Instead of manually scanning hundreds of text files, researchers can now ask direct questions and receive concise, context-aware responses.
The originality of this project lies in its focus on biomedical content and its accessible implementation. Unlike generic RAG demos, it integrates a clean Streamlit chat interface with ChromaDB, making it approachable for researchers, students, and developers who want to explore AI-driven literature analysis.
Conversation Memory
To enhance user experience, the assistant includes a small memory context.
This means that it can remember the most recent turns in the conversation, allowing for smoother and more coherent interactions.
The memory is intentionally kept lightweight to balance usability and efficiency, ensuring the system remains fast and resource-friendly.
Methodology
The system follows a modular pipeline:
Document Loading
Users can place .txt files containing cancer-related research into the /documents directory. These files serve as the knowledge base.
Chunking and Embedding
Documents are split into smaller segments (chunks) to improve retrieval accuracy. Each chunk is embedded into vector representations using a transformer model.
Vector Database with ChromaDB
The embeddings are stored in ChromaDB, enabling efficient similarity search. When a query is made, the database retrieves the most relevant chunks.
Retrieval-Augmented Generation
Retrieved chunks are passed as context to a large language model (LLM) through the Groq API. The chosen model, LLaMA 3.1 (8B Instant), generates a natural language response that integrates both retrieved knowledge and general reasoning ability.
Streamlit Chat Interface
A user-friendly chat page enables interaction. The system maintains conversation history, showing both user queries and AI-generated responses in sequence, replicating a familiar chat experience.
(diagram suggestion: flow from Documents → ChromaDB → LLM → Streamlit Chat)
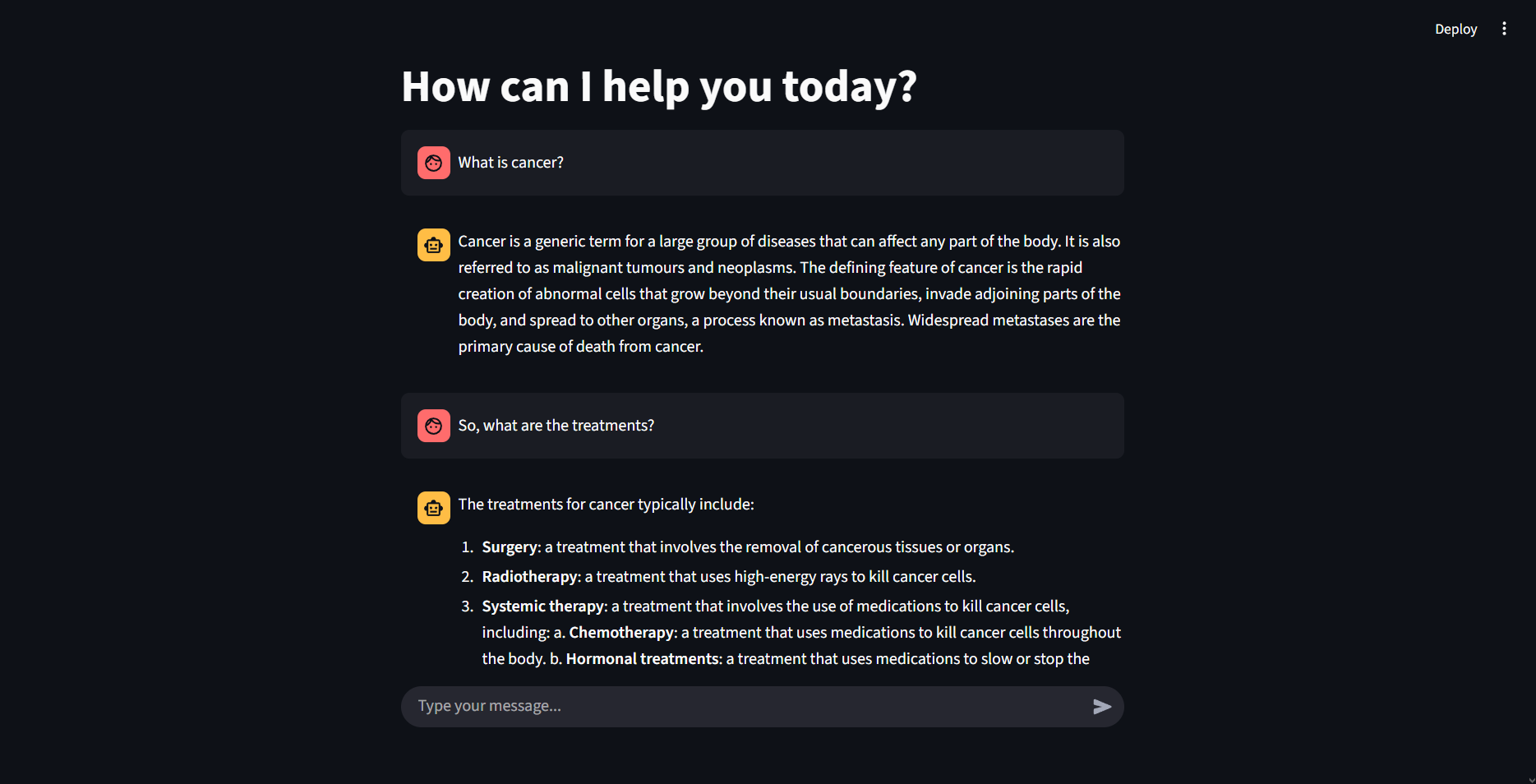
Results and Demonstration
The application provides a simple yet effective interface for biomedical exploration.
Example workflow:
A researcher loads a set of cancer-related .txt studies into /documents.
In the chat interface, the researcher asks:
Q: What are common treatments for lung cancer?
A: The assistant retrieves relevant chunks from the dataset, then generates a concise response outlining chemotherapy, targeted therapy, and immunotherapy as mentioned in the uploaded texts.
Another query:
Q: Summarize the risk factors highlighted in the dataset.
A: The system identifies and lists smoking, family history, and environmental exposure, as supported by the documents.
Importantly, the assistant is designed to decline answering when sufficient context is not found, ensuring transparency and reliability.
Example Prompts
To help first-time users, here are some sample questions you can try with your cancer research documents:
What are the main causes of breast cancer mentioned in the dataset?
Summarize the latest treatments described in the documents.
What are the most common side effects of chemotherapy according to the research?
Explain how genetics influence cancer risk in the loaded studies.
Which preventive strategies are highlighted across the documents?
List the biomarkers mentioned for early detection of lung cancer.
These examples show the flexibility of the assistant, allowing you to ask specific, high-level, or exploratory questions depending on your research needs.
Visual Demo
Conclusion
This project provides a lightweight, open-source framework for building RAG assistants tailored to cancer research. By integrating ChromaDB for retrieval, Groq-powered LLMs for reasoning, and Streamlit for accessibility, it showcases how AI can be applied in biomedical literature analysis.
With further development, this system can evolve into a more advanced tool supporting researchers, students, and practitioners. In its current form, it demonstrates the core principles of clarity, completeness, relevance, and engagement — aligning with Ready Tensor’s standards for impactful project publications.
License
This project is licensed under the MIT License — you are free to use, modify, and distribute it with attribution.