Abstract
BioMASS is an innovative multi-agent spatial model designed to enhance computational efficiency in simulations involving complex sensory and locomotion functions. Traditional agent-based modeling (ABM) platforms suffer from performance degradation as the number of agents and their perception ranges increase, leading to quadratic computational costs. BioMASS addresses this issue by employing a quadruply linked list structure, which enables constant-time neighborhood search and movement. It is implemented within a multi-agent system framework that has been successfully applied to marine ecosystem simulations, demonstrating its ability to track species interactions across multiple trophic levels in real-time, outperforming existing platforms.
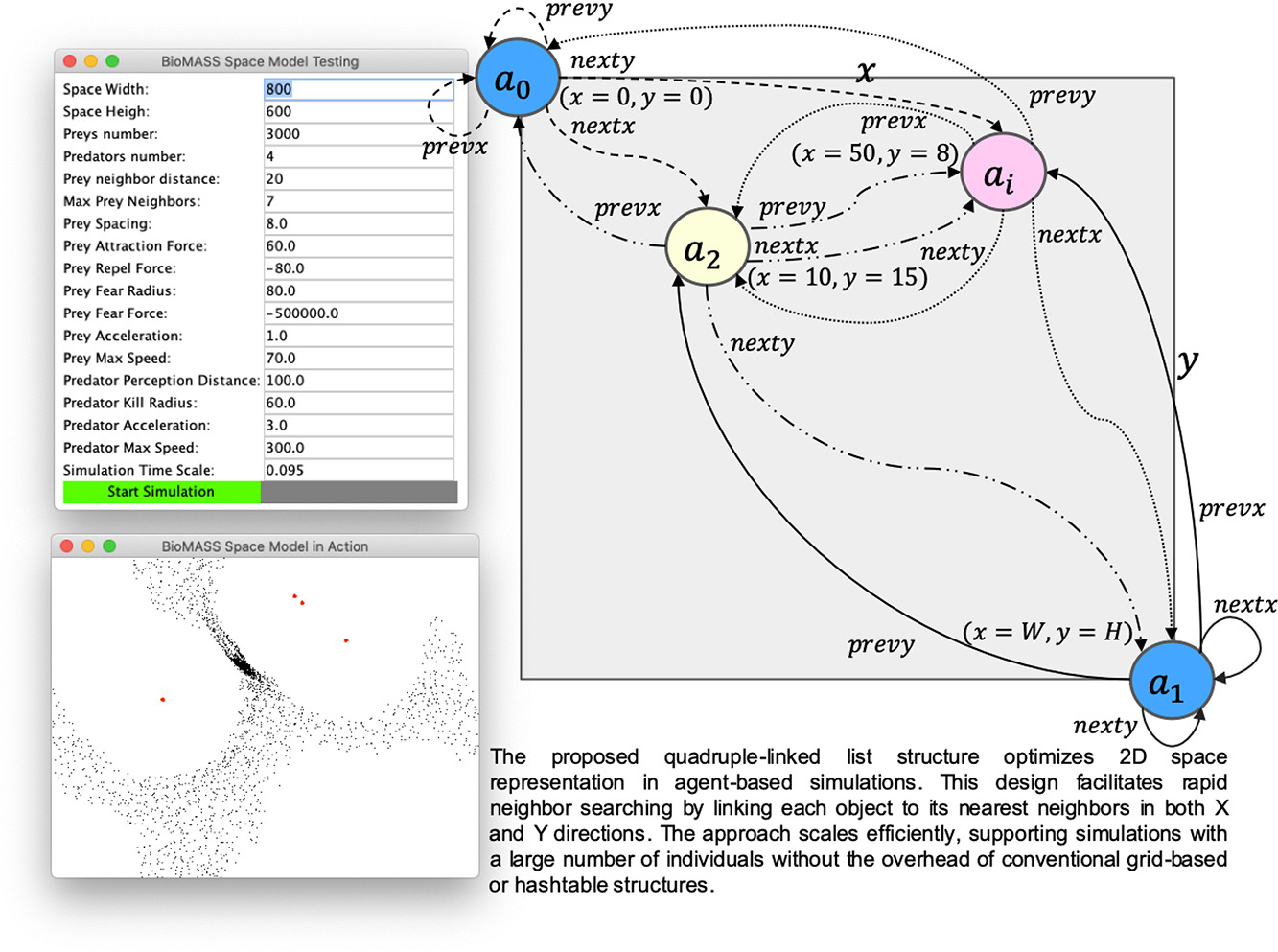
Fig.1. Graphical Abstract of BioMASS Model. The proposed quadruply linked list structure optimizes 2D space representation in agent-based simulations. This design enables efficient neighborhood search by linking each agent to its nearest neighbors in both X and Y directions. The approach scales effectively, supporting large-scale simulations without the computational overhead of grid-based or hash table structures.
Introduction
Agent-based modeling and simulation (ABMS) is a method used to study the dynamics of complex systems. It is widely applied across disciplines such as economics, ecology, sociology, and biology to investigate how interactions among individual components lead to self-organizing behaviors and emergent properties in complex systems [1, 2]. ABMS systems are designed as multi-agent systems (MAS) to distribute task-solving across agents. Within ABMS, two main types of models are used to study complex systems: behavioral simulations and agent-based system simulations, each suited to different experimental needs due to their distinct spatial and temporal resolutions.
In behavioral simulations, individual-level rules generate collective patterns and relatively few agents (typically tens to hundreds) are required. These simulations emphasize high spatial and temporal granularity, allowing researchers to observe how individual interactions contribute to group behaviors. Since the focus is on immediate interactions within small populations, these models do not require large spatial or temporal dimensions.
Agent-based system simulations, on the other hand, serve as complementary tools to traditional state-variable models, emphasizing physiological processes and local interactions that give rise to emergent behaviors at the population level. This paradigm requires a much larger number of agents (often thousands) and operates in expansive environments over extended time-frames, often spanning years or decades. The goal is to capture the evolution of population dynamics over multiple generations, requiring less granular temporal and spatial resolution compared to behavioral simulations.
Certain scenarios require blending both paradigms, especially when individual behaviors directly impact macro-level variables. These hybrid models add computational demands, as they need to manage large agent populations while accounting for individual variability in interactions, life cycles, and behaviors. Since these simulations are spatially explicit, agents interact primarily through sensory and perception functions, which adds computational complexity. Agents must frequently sense a large portion of their environment, requiring costly neighborhood search functions. Furthermore, variability in agents’ perception ranges increases the difficulty of optimizing these functions within traditional spatial models. This highlighted the need for a spatially explicit ABMS framework capable of efficiently handling neighborhood exploration and movement functions, addressing the computational challenges posed by large, spatially complex environments.
The Problem
The classical method adopted to solve these types of problems involves a discrete virtual space (usually a two- or three-dimensional rectangular lattice) and a search algorithm whose computational cost per individual increases with the perception range and with the number of discrete cells in the search area. To reduce this cost, the number of cells must be kept low. This is done either by reducing the perception range or by increasing cell size (sacrificing detail in the environment representation). Another approach is to use a continuous space where the computational cost per agent increases linearly with the number of agents. However, models with a large number of agents (1000s or more) perform poorly under this scheme. To avoid very costly processing in this case, a collection of agents can be modeled as a group and represented as a special kind of agent. But this again sacrifices detail in the model representation, especially regarding the degree of heterogeneity among the agents.
Oversimplifying the spatial representation or the number of individuals may work for certain systems. However, in [4], Sansores et al. described several scenarios that require large spatial dimensions, fine spatial granularity and a large number of agents at the same time. Moreover, the perception range of the agents may change over time, so it is desirable to preserve the heterogeneity that forms the foundation of ABMS models. The combination of these requirements creates an additional level of complexity in the spatial model that is not addressed by any current ABMS framework. Our main contribution focus on the BioMASS spatial model.
Classical spatial models for situated multi-agent systems
Traditionally, the internal model of the environment in a situated MAS is either discrete or continuous. This choice determines how the agents can be physically organized and the forms of the sensory and locomotion functions used by the agents. The hybrid model, a combination of the above, uses partial discretization of the underlying continuous model in order to improve the execution performance of some functions such as neighborhood search.
The Need for an optimized spatial model
Existing spatial models, see Fig.2, Fig.3, Fig.4, suffer from high computational costs:
- Discrete models rely on fixed grids, leading to inefficient neighborhood searches.
- Continuous models require global searches, making them computationally expensive.
- Hybrid models balance these approaches but require predefined discretization, limiting flexibility.
Research Questions
- What is the theoretical computational complexity of the BioMASS model in terms of time, and how does it compare to that of other spatial models used in agent simulations?
- To what extent does the BioMASS model reduce the computational complexity of neighborhood search and other spatial operations in agent simulations, especially in scenarios with a large number of agents and varying perception ranges?
- How can the BioMASS model be integrated into an existing agent simulation framework, and what are the benefits of this integration in terms of performance and scalability?
- To what extent does the BioMASS model facilitate the design and implementation of more efficient and scalable agent simulations for the study of complex systems?
Objectives
- Conduct a formal analysis of the computational complexity of the BioMASS model, deriving mathematical expressions for the time required by its main operations (insertion, removal, movement, neighborhood search).
- Compare the theoretical computational complexity of the BioMASS model with that of other spatial models (continuous, discrete, and hybrid) by analyzing their respective complexity expressions.
- Implement the BioMASS model in an open-source agent simulation framework and evaluate its performance in different simulation scenarios, measuring execution time and scalability.
- Evaluate the impact of the BioMASS model on the performance and scalability of a specific agent-based simulation application, such as the predator-prey model described in the article.
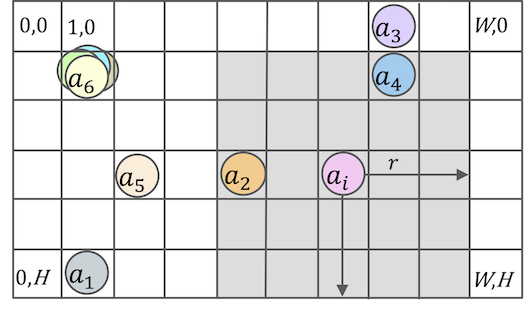
Fig.2. A discrete spatial model
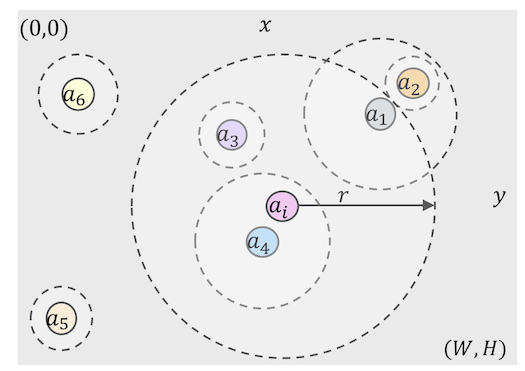
Fig.3. A continuous spatial model with Cartesian coordinates. The set of agents
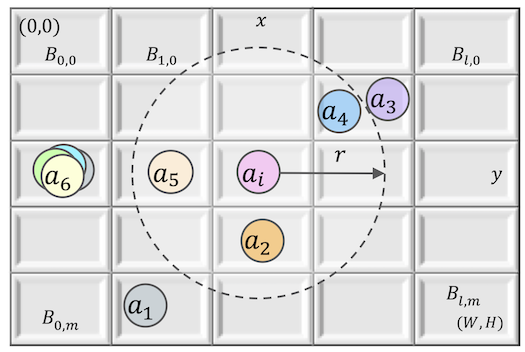
Fig.4. A hybrid spatial model with a continuous space divided into buckets. The set of agents
Methodology
The proposed BioMASS quadruply linked list spatial model
Let
The initial setup is illustrated in Fig.5. The sentinel node
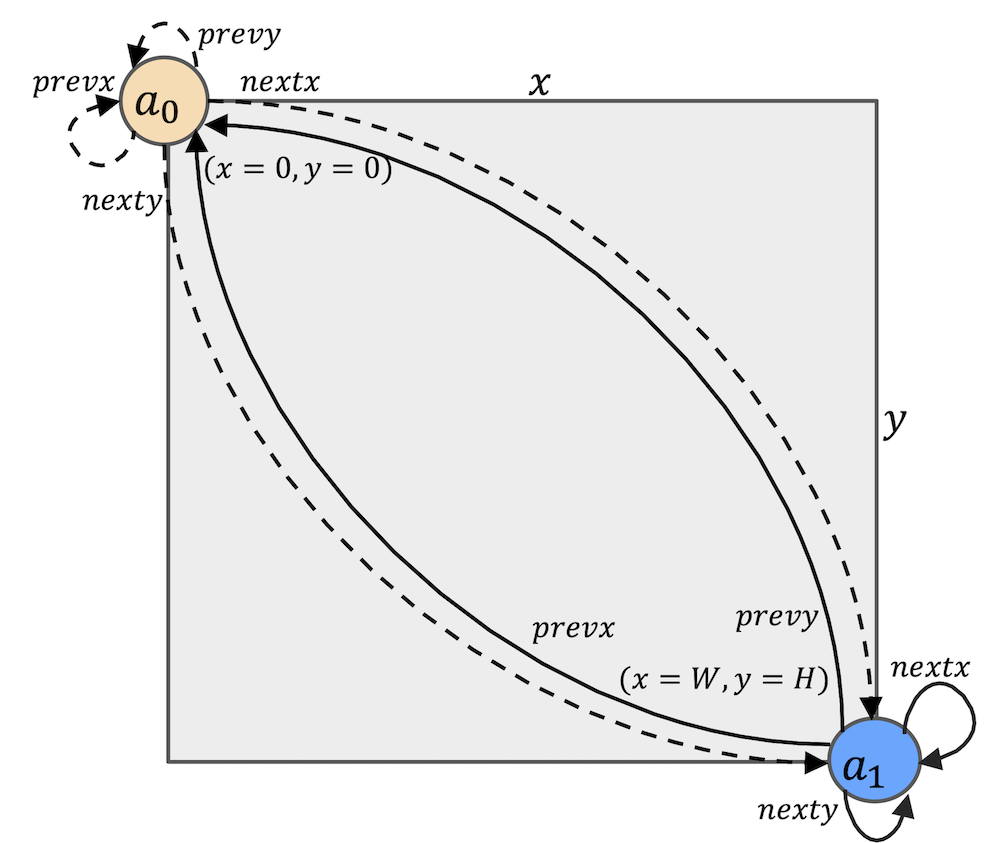
Fig.5. The quadruply linked list in
Fig.6 and Fig.7 illustrate the insert function. The time complexity of this function is
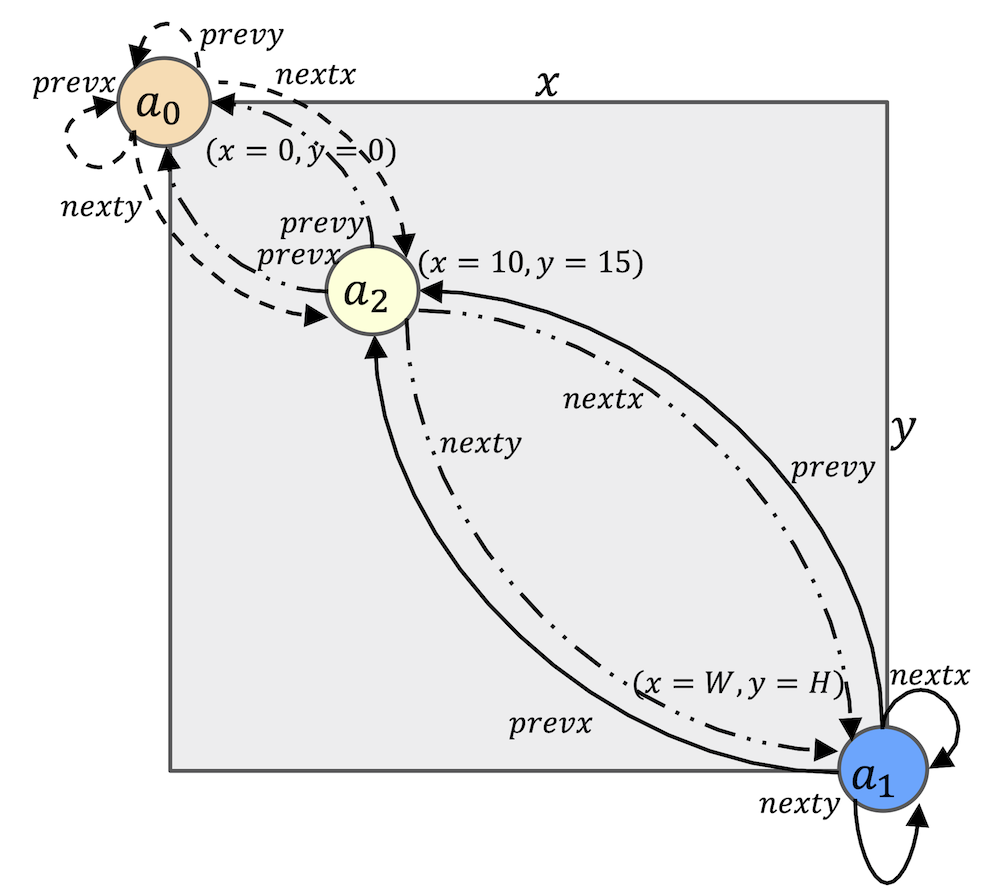
Fig.6. The space
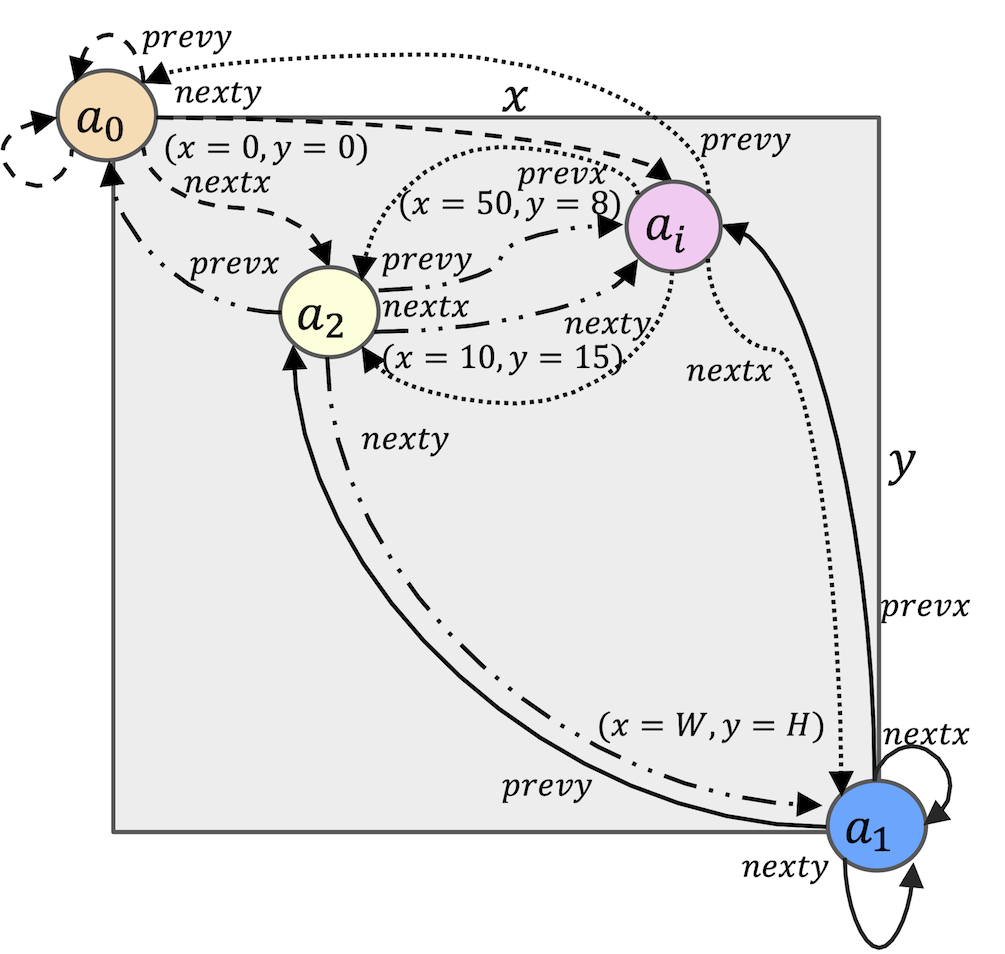
Fig.7. The space
The neighborhood lookup function
Let
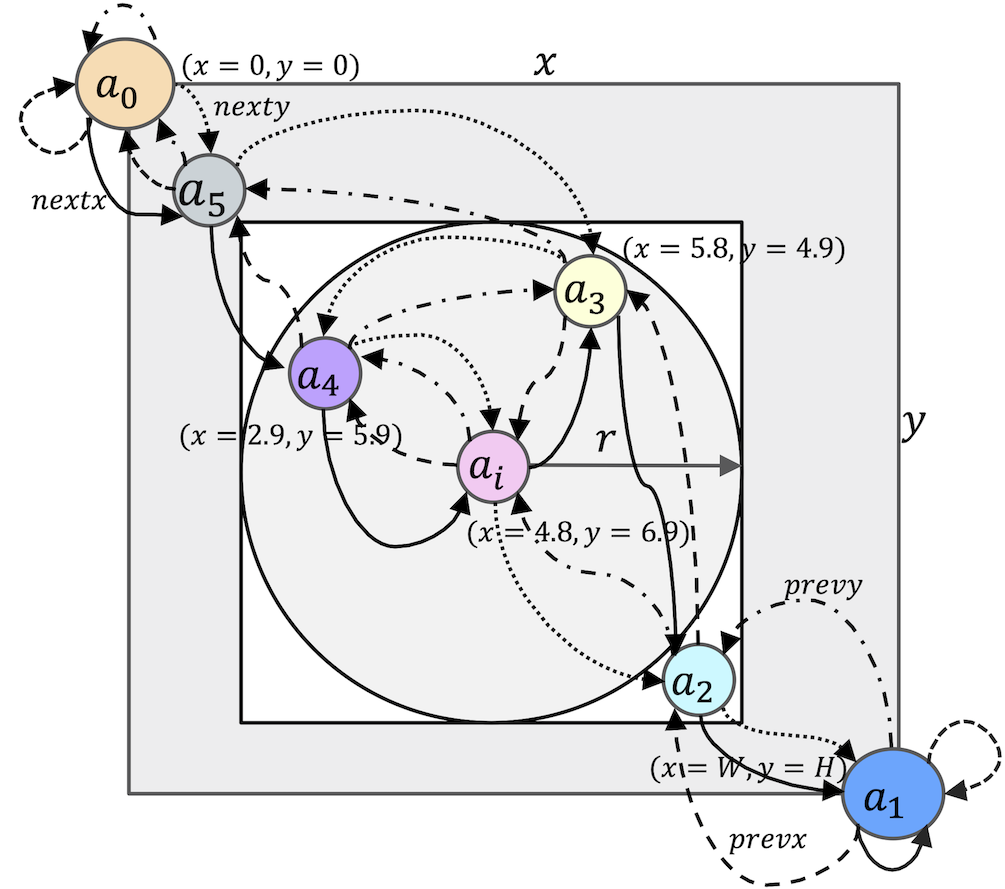
Fig.8. BioMASS spatial model depicting the neighborhood
The neighborhood lookup algorithm
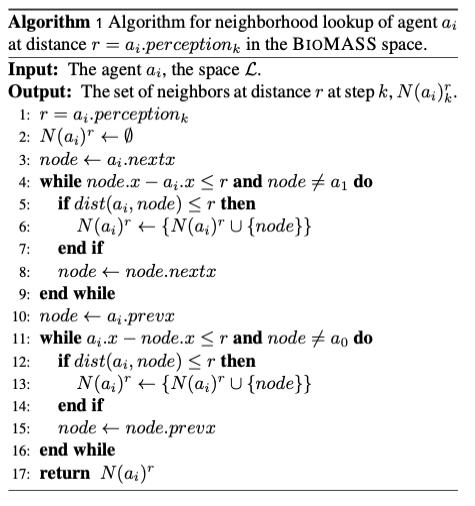
Fig.9. Algorithm for neighborhood lookup of agent
Complexity analysis: strengths and limitations
For theoretical analysis and detailed algorithmic evaluation, see Sansores et al., 2020.
Table 1 summarizes the time complexities of the different spatial models. BioMASS has the best theoretical performance with respect to the time complexity of the neighborhood lookup function, but the time complexity of its insert function is high compared to the other models. However, most simulation experiments are not limited by the insertion time of agents, since this is done just once to set up the initial scenario. Rather, to model the dynamics of complex systems with local interactions, the sensory function is of primary importance since it is used most intensively. In the case of the BioMASS space, the time complexities of the move and neighborhood lookup functions are both
The worst case implies that the density of agents is high compared to the perception scope. Such cases are rare in the simulation of complex systems. When they arise, it often means that the resolution of the simulation is unsuitable for the problem. Furthermore, if they do happen frequently, the functions can be implemented differently. For example, if agents are able to perceive the whole space then it is simpler to provide them with a list of all individuals in the space. In contrast, in the continuous space the neighborhood lookup function is always
| Operation | Discrete Model | Continuous Model | Hybrid Model | BioMASS |
|---|---|---|---|---|
| Insertion | O(1) | O(1) | O(1) | O(n) |
| Deletion | O(1) | O(1) | O(n) | O(1) |
| Movement | O(1) | O(1) | O(n) | O(1) |
| Neighborhood Lookup | O(W²) | O(n) | O(n × W²) | O(1) |
*with W = width of the space
Experiments
Performance benchmarking
We compare the four spatial models through a case study: the classical swarm model. The case study simulates a large flock of prey birds that attempt to avoid a small number of predator birds flying in the same environment. Individual prey and predator birds are modeled as autonomous agents living in a 2D toroidal (opposite boundaries of the space are joined) continuous space.
Since the BioMASS spatial model implements a continuous space with Cartesian coordinates, we decided to compare it only to similar models. That is, we did not test the swarm scenario with the discrete spatial model, which performs poorly in large spaces with neighborhood lookup ranges beyond the agent's surrounding cells.
We selected two simulation platforms for comparison, Repast and MASON. Our selection criteria were as follows: (1) the platform supports one of the two models with Cartesian coordinates, continuous or hybrid; (2) it is a general-purpose simulation tool; and (3) it is an ongoing project still releasing new versions. Also, the Java programming language was an important requirement to standardize the experiments between simulation platforms. Finally, while selecting the platforms we also took into consideration less critical characteristics such as robustness, running speed, and open-source support.
All simulations were executed on an iMac with a 3.2 GHz Quad-Core Intel Core i5 processor. The iMac had 16GB of memory and was running MacOS Catalina.
The experiments measured execution time per simulation step, demonstrating that BioMASS consistently outperformed other models in both homogeneous and heterogeneous agent configurations.
Source code
The spatial model is integrated into a simple yet versatile Java framework for multi-agent system simulation. Key classes within this framework include Agent.java, Scheduler.java, Space.java and PhysicalObject.java, which coordinate the core functions of agent behavior, simulation timing, and spatial environment management. The PhysicalObject.java class specifically equips agents with the four pointers necessary for insertion into the quadruply linked list. The orchestration of a complete simulation can be observed in the source code and example available in the GitHub repository, where the Main.java class serves as the entry point. Executing this class launches a graphical user interface (GUI) that enables users to configure key parameters for the provided example, including the simulation space size, the number of agents, and the perception range. Notably, this framework is designed with flexibility in mind, allowing for easy adaptation to different scenarios beyond the example provided. This adaptability makes it straightforward for users to modify the framework for a variety of multi-agent simulations by adjusting or extending its core components.
Dataset
The BioMASS Space Model Dataset Benchmark
is publish in the IEEEDataPort to easily reproduce the experiments. This dataset contains the results of the simulation runs of the experiments performed to evaluate and compare the proposed spatial model for situated multi-agent systems.
Dataset processing
It is important to note that this research is based on data generated by simulations. Therefore, traditional dataset preprocessing methodologies are not applicable. The data used for analysis consists of the parameters used to configure the simulations, such as the number of agents, perception ranges and environment size, and the resulting performance metrics, such as time steps and execution times. The parameters of the simulations were varied in order to test the performance of the BioMASS model under a wide variety of conditions.
Instructions
The dataset include a compressed file in zip format. It contains a directory structure as shown below. Each directory is a specific experiment with each simulation toolkit and parameters. Inside each directory there are 50 CSV files, one for echa simulation run. Each file has a header describing the main parameters of the corresponding experiment. We use the Repast Toolkit, and Mason Toolkit to perform a benchmark with the proposed BioMASS spatial model.
Directory:
Experiments
-heterogeneous
heter-biomass
heter-mason.20
heter-mason.40
heter-mason.200
heter-mason.400
-homogeneous
homo-biomass
homo-mason.200
homo-mason.400
homo-repast
The simulation setup and full benchmarking methodology are described in Sansores et al., 2020.
Validation Approach
To validate the performance of the BioMASS model, an empirical validation approach was adopted, based on experimentation and comparison with other established simulation models. This approach focuses on evaluating the model's ability to accurately simulate the behavior of complex systems and its computational efficiency in various scenarios.
Verification methods
- Simulation replication: Multiple simulation runs (50 repetitions) were performed for each experimental scenario. This allowed us to assess the consistency and stability of the results obtained with the BioMASS model.
- Comparison with existing models: The performance of the BioMASS model was compared with that of other widely used spatial models, such as continuous (Repast) and hybrid (MASON) models. This comparison was based on key performance metrics, such as simulation runtime and model scalability.
- Theoretical Complexity Analysis: A formal analysis of the computational complexity of the BioMASS model was performed, deriving mathematical expressions for the time required by its main functions. This analysis was compared with the theoretical complexity of other spatial models to validate the efficiency of the BioMASS model from a theoretical perspective.
- The maximum, minimum, and average values of the time step were calculated for each simulation run. These values were used to assess the variability and stability of the model's performance across multiple runs.
Robustness and quality considerations
- Scenario variability: Various simulation scenarios were used with different numbers of agents, perception ranges, and environment sizes to evaluate the robustness of the BioMASS model under a wide range of conditions.
- Statistical analysis: Statistical analysis techniques were applied to the simulation results to assess the significance of the performance differences observed between the BioMASS model and other models.
- Results validation: The results obtained with the BioMASS model were compared with the expected results based on theory and empirical evidence available in the scientific literature.
Validation Results
The theoretical complexity analysis demonstrated that the BioMASS model has a lower computational complexity for the neighborhood search function compared to other models. This efficiency of the BioMASS model was confirmed by the empirical validation results, which showed that the BioMASS model outperforms traditional spatial models in terms of computational efficiency, especially in scenarios with a large number of agents and varying perception ranges.
Validation implications
The validation results support the validity and usefulness of the BioMASS model as an efficient and accurate tool for simulating complex systems. The BioMASS model has the potential to facilitate the study of a wide range of complex phenomena in various scientific disciplines.
Case study: marine ecosystem simulation
BioMASS was tested in a marine ecosystem simulation, where it successfully modeled predator-prey interactions, trophic chains, and adaptive behaviors. The simulation demonstrated BioMASS’s ability to:
- Track species interactions across multiple trophic levels.
- Maintain real-time execution even with thousands of agents.
- Handle dynamic perception ranges efficiently.
For a more detailed description of the marine ecosystem simulation, refer to Sansores et al., 2016.
Results
We analyzed the time complexity of the different spatial models experimentally through simulation. The simulation experiments tested various perception scopes, to test the execution time of the spatial model for extreme values. The results of these experiments are depicted in Figs. 10 to 15, and clearly demonstrate that BioMASS outperforms the other models.
Key Findings
The experimental results highlight:
- Scalability: BioMASS handles thousands of agents efficiently, unlike traditional models.
- Computational efficiency: O(1) complexity in neighborhood lookup, compared to O(n) or worse in other models.
- Support for heterogeneous agents: BioMASS successfully manages dynamic perception ranges without degrading performance.
- Comparison to classical models: BioMASS outperforms grid-based, continuous, and hybrid models in both speed and scalability. This approach eliminates the need for global searches, reducing the computational cost of spatial queries.
For an in-depth discussion of the experimental results and performance metrics, refer to Sansores et al., 2020 and Sansores et al., 2016.
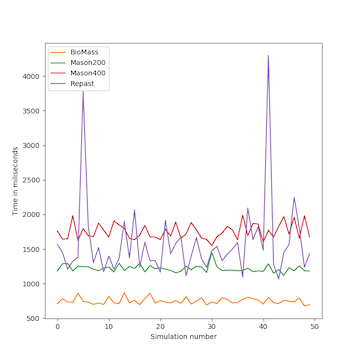
Fig.10. Maximum time step duration of each run for the simulations with homogeneous perception scope.
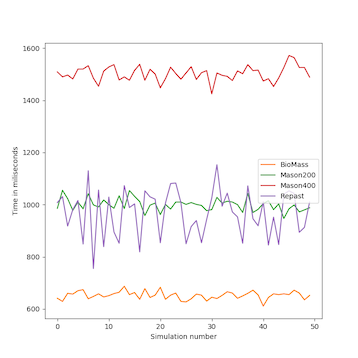
Fig.11. Minimum time step duration of each run for the simulations with homogeneous perception scope.
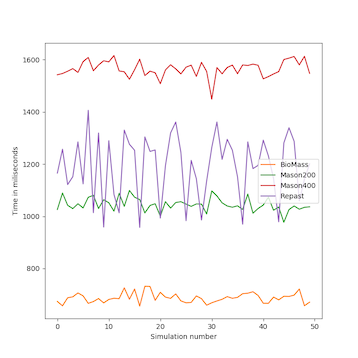
Fig.12. Average time step duration of each run for the simulations with homogeneous perception scope.
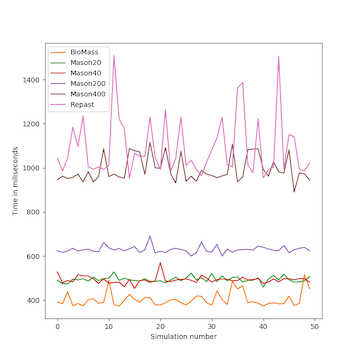
Fig.13. Maximum time step duration of each run for the simulations with heterogeneous perception scope.
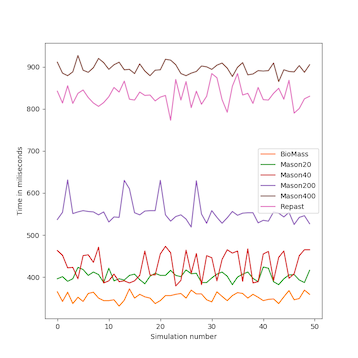
Fig.14. Minimum time step duration of each run for the simulations with heterogeneous perception scope.
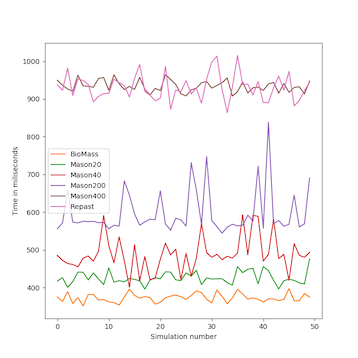
Fig.15. Average time step duration of each run for the simulations with heterogeneous perception scope.
Conclusion
The BioMASS space model represents a significant advancement in the field of multi-agent systems, offering researchers an efficient and scalable tool for simulating large-scale, spatially explicit environments. By addressing the computational inefficiencies present in traditional ABM platforms, BioMASS enables more detailed and realistic simulations of complex systems, such as marine ecosystems or predator–prey dynamics. The main reason for this accomplishment is that the spatial model is designed to perform the neighborhood search function with
of agents to use the search function intensively without significantly affecting the simulation time.
Future work
Future work could extend the model to support three-dimensional environments, further broadening its application.