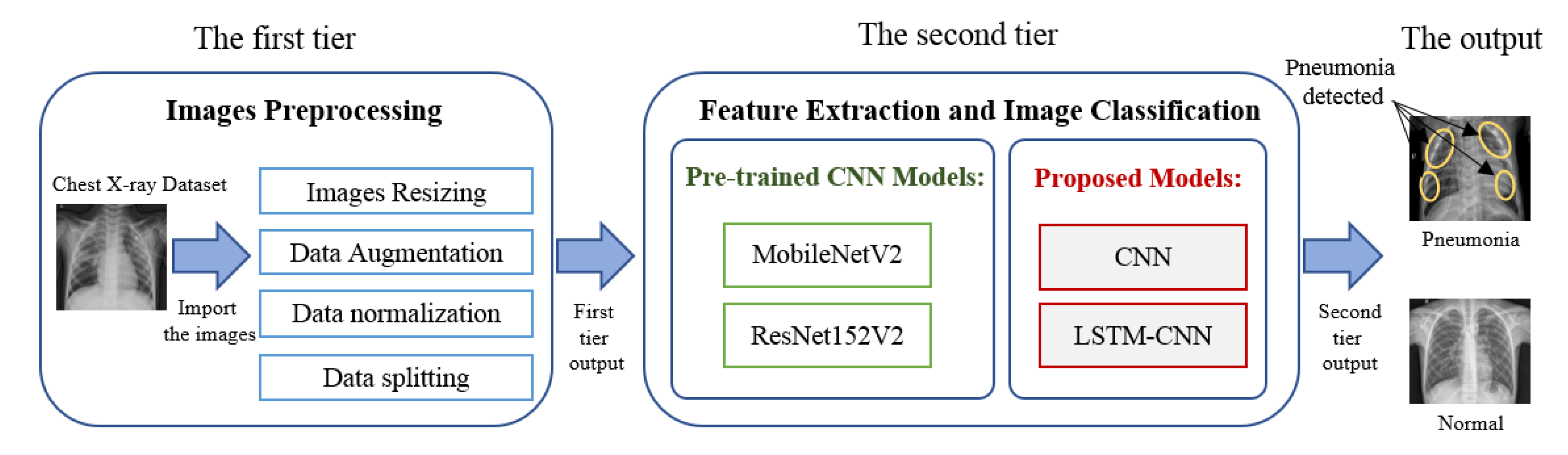
Project Documentation: Pneumonia Detection from Chest X-ray using CNN(Computer Vision) :
Github - https://github.com/DebansuprasadaSahoo/pneumonia-detection-using-computer-vision
Pneumonia: A Medical Overview :
Pneumonia is a respiratory infection that inflames the air sacs in the lungs (alveoli). It can be caused by bacteria, viruses, or fungi. Symptoms include coughing, fever, and difficulty breathing, and it can be life-threatening if not diagnosed and treated promptly.
LIVE LINK - https://pneumonia-detection-using-computer-vision-7bwihx2cab9dxmydvued.streamlit.app/
Challenges in Diagnosis:
- Diagnosing pneumonia through chest X-rays can be subjective and prone to human error.
- Radiologists must interpret numerous images daily, increasing the risk of oversight in busy clinical environments.
- Variations in imaging quality and the presentation of pneumonia in X-rays make diagnosis more complex
Importance of the Project :
-
Early Detection: Pneumonia is a leading cause of mortality, especially in children and the elderly. Early detection can drastically improve treatment outcomes.
-
Automation in Healthcare: The use of CNNs automates the diagnostic process, reducing dependency on manual interpretation by radiologists.
-
Scalability: Once deployed, the model can be scaled globally, making quality healthcare accessible in underserved regions.
-
Efficiency: The system drastically reduces the time and effort required to analyze X-rays manually.
-
Accuracy: Deep learning models often surpass traditional methods and even human experts in terms of diagnostic accuracy.
This project has the potential to revolutionize the medical industry by integrating AI with routine diagnostic workflows, making healthcare smarter, faster, and more reliable.
Convolutional Neural Networks (CNNs): Architecture and Functionality :

- Core Idea of CNNs
CNNs are inspired by the way the human brain processes visual information. The network learns to recognize patterns, such as edges, textures, and shapes, and progressively combines them to identify complex objects or abnormalities.
In this project, the CNN learns to detect pneumonia-specific patterns in chest X-rays.
- Key Layers in CNN Architecture
-
Convolutional Layers:
These layers detect features (e.g., edges, textures) by applying filters (kernels) over the input image.
Example: A filter might identify the boundary between normal and inflamed lung regions. -
Activation Functions (ReLU):
The Rectified Linear Unit (ReLU) introduces non-linearity, enabling the network to learn complex patterns.
After convolution, ReLU ensures only significant activations (positive values) are passed forward. -
Pooling Layers (MaxPooling):
Pooling reduces the spatial dimensions of the feature maps, making computation efficient and focusing on the most critical features.
In this project, pooling layers ensure the model generalizes well to unseen X-rays. -
Flattening:
The output of the convolutional and pooling layers is flattened into a 1D array, preparing it for fully connected layers.
-nFully Connected Layers:
These layers combine all detected features to make the final classification (pneumonia or normal).
- Output Layer with Sigmoid Activation:
The sigmoid activation outputs a probability score (e.g., 0.8 means an 80% chance of pneumonia).
- Training the Model
-
Forward Propagation:
Input X-ray images pass through the layers, generating predictions. -
Loss Calculation:
The binary cross-entropy loss compares predictions to actual labels, measuring the model's performance. -
Backward Propagation:
Gradients of the loss with respect to each weight are computed, enabling the model to adjust its parameters using optimization techniques like Adam.
Dataset Overview :
Dataset - https://www.kaggle.com/datasets/paultimothymooney/chest-xray-pneumonia
The dataset used for this project consists of labeled chest X-ray images sourced from publicly available repositories like Kaggle or medical
imaging archives. These datasets typically categorize images into two classes:
- Normal: Chest X-rays without any signs of pneumonia.
- Pneumonia: Chest X-rays showing signs of bacterial or viral pneumonia.
The dataset is split into training, validation, and testing subsets to ensure the model's performance is robust and generalizable.
Structure :
The dataset is structured into separate folders for training and testing data. Each folder contains subfolders for each class (e.g., 'Normal' and 'Pneumonia'):
/Normal
image1.jpg
image2.jpg
...
/Pneumonia
image1.jpg
image2.jpg
...
/test
/Normal
image1.jpg
image2.jpg
...
/Pneumonia
image1.jpg
image2.jpg
...
Backend Code Explanation :
- Data Preparation
import os from tensorflow.keras.preprocessing.image import ImageDataGenerator # Specify dataset paths train_data_path = 'path_to_train_data' test_data_path = 'path_to_test_data' # Check if the dataset paths exist and have the correct structure if not os.path.exists(train_data_path): raise FileNotFoundError(f"Training data path not found: {train_data_path}") if not os.path.exists(test_data_path): raise FileNotFoundError(f"Testing data path not found: {test_data_path}") # Ensure that the directory structure is correct if not any(os.listdir(train_data_path)): raise FileNotFoundError("Training data directory is empty or improperly structured.") if not any(os.listdir(test_data_path)): raise FileNotFoundError("Testing data directory is empty or improperly structured.")
- Data Augmentation
# Data augmentation for training data datagen = ImageDataGenerator( rescale=1.0/255, rotation_range=30, width_shift_range=0.1, height_shift_range=0.1, shear_range=0.2, zoom_range=0.2, horizontal_flip=True ) train_data = datagen.flow_from_directory( train_data_path, target_size=(224, 224), batch_size=32, class_mode='binary' ) # Data preprocessing for testing data test_datagen = ImageDataGenerator(rescale=1.0/255) test_data = test_datagen.flow_from_directory( test_data_path, target_size=(224, 224), batch_size=32, class_mode='binary' )``` 3. Model Architecture ```python from tensorflow.keras.models import Sequential from tensorflow.keras.layers import Conv2D, MaxPooling2D, Flatten, Dense from tensorflow.keras.optimizers import Adam # Build the CNN model model = Sequential([ Conv2D(32, (3, 3), activation='relu', input_shape=(224, 224, 3)), MaxPooling2D(pool_size=(2, 2)), Flatten(), Dense(128, activation='relu'), Dense(1, activation='sigmoid') # Binary output ]) model.compile(optimizer=Adam(), loss='binary_crossentropy', metrics=['accuracy'])
- Training and Evaluation
# Train the model model.fit(train_data, validation_data=test_data, epochs=10) # Save the model for later use model.save('pneumonia_model.h5')
- Confusion Matrix Visualization
from sklearn.metrics import confusion_matrix import matplotlib.pyplot as plt # Evaluate the model predictions = (model.predict(test_data) > 0.5).astype(int) cm = confusion_matrix(test_data.classes, predictions) # Plot the confusion matrix plt.matshow(cm, cmap='Blues') plt.title("Confusion Matrix") plt.colorbar() plt.xlabel("Predicted") plt.ylabel("True") plt.show()
Frontend Code Explanation :
- File Upload and Preprocessing
# Import required libraries import streamlit as st from PIL import Image import numpy as np from tensorflow.keras.models import load_model # File upload interface uploaded_file = st.file_uploader("Choose an X-ray image...", type=["jpg", "png", "jpeg"]) if uploaded_file is not None: image = Image.open(uploaded_file) st.image(image, caption='Uploaded X-ray', use_column_width=True) # Convert the image to RGB (if not already in RGB format) image = image.convert("RGB") # Preprocess the image image = image.resize((224, 224)) image = np.array(image) / 255.0 # Normalize the pixel values to [0, 1] image = np.expand_dims(image, axis=0) # Add batch dimension
- Model Integration
# Load the trained model model = load_model('pneumonia_model.h5') # Generate prediction prediction = model.predict(image) # Display the prediction if prediction > 0.5: st.write("Prediction: Pneumonia Detected") else: st.write("Prediction: Healthy")
- Styling
# Add custom CSS for background image bg_image_url = "https://wallpaperbat.com/img/304560-doctor-wallpaper.jpg" # Replace with your image URL st.markdown( f""" <style> .stApp {{ background-image: url("{bg_image_url}"); background-size: cover; background-position: center center; background-repeat: no-repeat; height: 100vh; }} </style> """, unsafe_allow_html=True
frontend image :

Some of the images of its working,
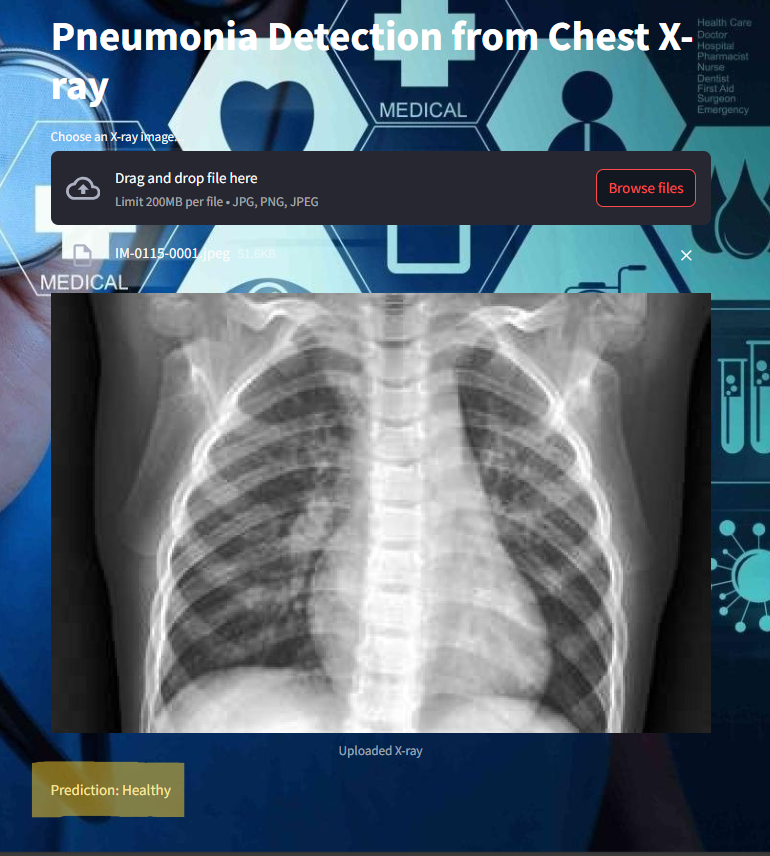
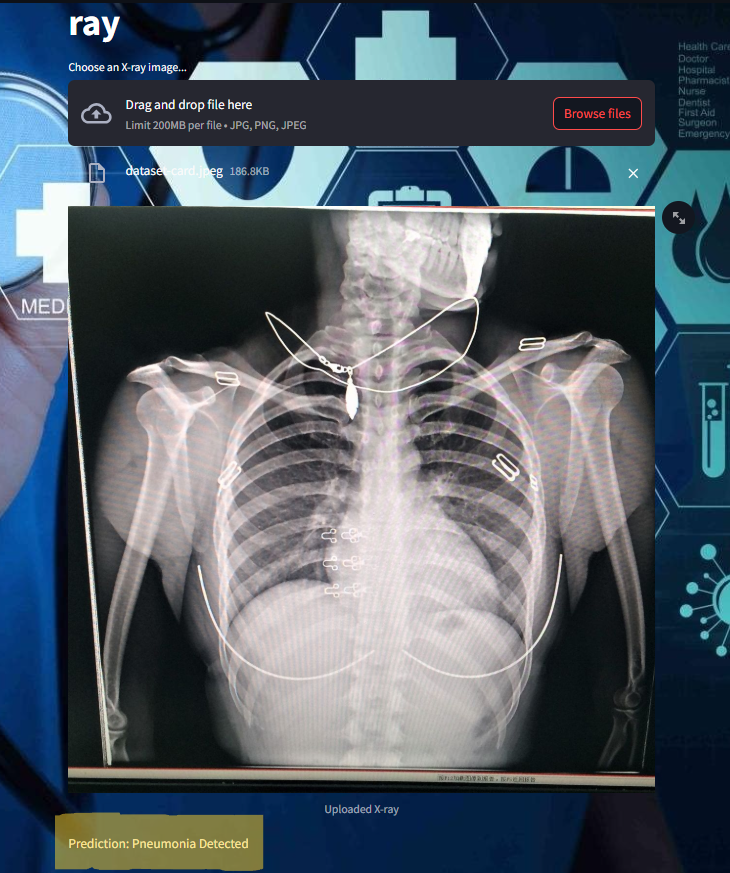
How This Project Revolutionizes the Medical Industry
1. Addressing Diagnostic Challenges
- Problem: Pneumonia diagnosis often requires expert radiologists to interpret X-ray images, which can be subjective and time-consuming.
- Solution: This AI-based approach offers consistent and objective analysis, minimizing errors due to human fatigue or oversight.
2. Scalability in Low-Resource Areas
- In underdeveloped or rural areas, access to trained radiologists is limited. By deploying the model in healthcare systems, even non-experts can use the application for preliminary screenings.
3. Reducing Turnaround Time
- Manual interpretation of X-rays can take hours or days in busy hospitals. The model delivers results in seconds, allowing for immediate action.
4. Enhancing Radiologist Efficiency
- Instead of replacing radiologists, this tool acts as a support system, flagging potential cases for further review. This hybrid approach ensures faster and more accurate diagnostics.
5. Cost-Effective Diagnosis
- Training radiologists and maintaining diagnostic infrastructure can be expensive. AI-driven solutions are a one-time investment, reducing long-term costs for healthcare providers.
6. Global Standardization
- The model learns from large, diverse datasets, ensuring that its predictions align with international diagnostic standards. This reduces regional variations in diagnostic accuracy.
7. AI-Powered Research
- The collected diagnostic data can be analyzed to study pneumonia trends, enabling proactive measures like vaccination drives or resource allocation during outbreaks.
By integrating AI into diagnostic workflows, this project not only improves patient outcomes but also enhances the overall efficiency and accessibility of healthcare systems. It demonstrates how deep learning technologies can bridge the gap between cutting-edge research and practical applications in medicine.