
Welcome to our publication on integrating hyperparameter tuning into our reusable machine learning models. In this publication, we'll leverage Optuna to optimize the performance of our existing Random Forest time step classifier model.
Here's a brief outline of what we'll cover:
By the end of this tutorial, you'll have a solid understanding of how to enhance your machine learning model's performance through hyperparameter tuning, adding another layer of adaptability to your model implementations. Let's get started!
Code Repository
The concepts in this publication are implemented in this repository:
https://github.com/readytensor/rt_tspc_random_forest_sklearn
In the world of machine learning, hyperparameters critically influence a model's performance. These are parameters set before the learning process starts, and finding the right combination can be a complex task. This is where hyperparameter tuning comes in, with several approaches available, each with their pros and cons. These include manual search, grid search, and random search.
Manual search, as the name suggests, involves manually adjusting hyperparameters and observing the results. This approach is highly time-consuming and requires a solid understanding of how different hyperparameters affect the learning algorithm.
Grid search and random search are more systematic. Grid search involves defining a set of possible values for each hyperparameter and systematically testing all combinations. This method is exhaustive and guarantees finding the best combination within the defined grid but can be computationally expensive. On the other hand, random search randomly selects combinations of hyperparameters to test, which can be more efficient but may miss the optimal combination.
In this tutorial, we opt for a more sophisticated method: Bayesian optimization using Gaussian Processes, implemented through the Optuna library. Bayesian optimization provides a principled technique based on information from past evaluations to find the minimum of a function. We chose Bayesian optimization because it can find better results with fewer function evaluations compared to grid search and random search. It's more suitable for high-dimensional hyperparameter spaces and scenarios where function evaluations are costly, which is typical in machine learning model training.
By the end of this tutorial, you'll understand how to perform hyperparameter tuning using Optuna and integrate it into your machine learning projects for better model performance. Let's dive in!
Hyperparameter tuning is a process in machine learning that essentially boils down to an optimization problem. Here, we're attempting to discover the ideal hyperparameter values that maximize our model's performance. The objective function, which we aim to minimize, in this context is our model's validation error. The variables we manipulate to achieve this are the hyperparameters. The goal, therefore, is to find those hyperparameter values that lead to the minimum validation error.
However, this process isn't straightforward. Training machine learning models is a resource-intensive task that requires significant computational power and time, particularly when dealing with models that have numerous hyperparameters. Given these constraints, it's desirable to reduce the number of objective function evaluations while still arriving at a satisfactory solution. This is precisely where Bayesian Optimization becomes invaluable.
In essence, Bayesian Optimization is designed to locate the minimum of a function with as few iterations as possible. It works by constructing a posterior distribution of functions, typically using Gaussian processes, that serves as the best approximation of the function we're trying to optimize. As we gather more observations, this posterior distribution becomes increasingly refined, aiding the model in determining which regions of the hyperparameter space are worth exploring versus those that aren't.
Thus, the optimization process is a delicate balancing act between exploration, where we sample areas of high uncertainty, and exploitation, where we sample areas estimated to have a good performance. In practical terms, this means we strive to locate the optimal values with the fewest steps possible. This efficiency is achieved by employing a surrogate optimization problem - in this case, finding the maximum of the acquisition function - which is substantially cheaper to evaluate compared to the original optimization problem.
Bayesian Optimization involves the following two core components:
1 - Surrogate model
2 - Acquisition function
Let's review each of these components in more detail.
The surrogate model plays a crucial role in Bayesian optimization and it's important to understand it.
A surrogate model is an approximation of the objective function that is cheaper to evaluate. It is built based on the evaluations of the objective function at previously sampled points. The surrogate model not only provides an estimate of the objective function at any point in the hyperparameter space but also quantifies the uncertainty of this estimate. Areas of the hyperparameter space that have not been sampled much will have higher uncertainty.
In Bayesian optimization, a Gaussian Process is often used as the surrogate model because it provides a measure of uncertainty for the function estimate. Other surrogate models include Random Forests and Gradient Boosting Machines.
The surrogate model is used to select the next point to evaluate in the hyperparameter space. This is done through the acquisition function, which takes into account both the estimate of the objective function from the surrogate model and the associated uncertainty.
In conclusion, the surrogate model is a critical component of the Bayesian optimization process. It provides a balance between exploration (sampling in areas of high uncertainty) and exploitation (sampling in areas estimated to have good performance) that is key to the efficiency of Bayesian optimization.
The acquisition function is used by the Bayesian optimization process to decide where to sample next. It trades off exploitation (sampling where the surrogate model predicts a good objective) and exploration (sampling in areas of high uncertainty). We will be using the Expected Improvement (EI) function which is a commonly used function in Bayesian optimization, including in hyperparameter tuning.
The EI function provides a balance between exploration and exploitation. At each step, the point with the highest expected improvement is chosen as the next point to evaluate.
In simple terms, EI gives a score to every point in the hyperparameter space. This score is high if the point is expected to improve upon the current best hyperparameter configuration (exploitation), and also if the uncertainty at that point is high (exploration). Thus, EI helps to guide the search process towards potentially optimal areas.
Besides Expected Improvement, there are other acquisition functions you could use:
Probability of Improvement (PI): This function selects the next point where the probability of improving upon the best observed point so far is the highest. It tends to focus more on exploitation rather than exploration.
Lower Confidence Bound (LCB): This function chooses the point that has the lowest value of the surrogate model minus a constant times the uncertainty. By adjusting the constant, we can control the balance between exploration and exploitation.
Upper Confidence Bound (UCB): Similar to LCB, but it chooses the point that has the highest value of the surrogate model plus a constant times the uncertainty. It is used when we want to maximize the objective function.
Entropy Search (ES) and Predictive Entropy Search (PES): These are more complex acquisition functions that aim to reduce the entropy of the distribution over the minimum of the objective function.
Knowledge Gradient (KG): This function estimates the improvement in the optimal solution resulting from the addition of a sample at a specific location.
The choice of acquisition function depends on your specific problem and computational resources, as some acquisition functions are more computationally demanding than others.
Let us now proceed to implementing hyperparameter tuning using Optuna.
Optuna, is a Python library that specializes in optimization tasks. We will use it for tuning the hyperparameters for our random forest classifier.
To install Optuna, you can use pip, a package installer for Python. Open your command line and run the following command:
pip install optuna
Optuna vs. Other Libraries
While Optuna is an excellent tool for hyperparameter tuning machine learning models, it's worth noting that it isn't the only one. Other libraries such as Hyperopt, Scikit-Optimize, and Spearmint also offer robust hyperparameter tuning capabilities. Each of these libraries comes with their unique features and advantages, and the choice of library often comes down to specific project requirements and personal preference.
Optuna offers a few methods for hyperparameter tuning, including Grid Search, Random Search, and Bayesian Optimization. In this tutorial, we'll focus on Bayesian Optimization.
To execute hyperparameter tuning using Bayesian optimization in Optuna, we require three main components:
1- A function to optimize: This is typically the model's performance evaluated on a validation set with a given set of hyperparameters.
2- A search space: This is a predefined range of potential hyperparameter values within which we are interested in searching.
3- An sampler: This refers to the particular optimization method used to search through the hyperparameter space. In Optuna, one commonly used sampler is TPE Sampler.
In a machine learning context, the function to optimize is often a wrapper around your model's training and evaluation procedure. This function should take as input a set of hyperparameters, train your model with these hyperparameters, evaluate it on a validation set, and then return the evaluation metric you wish to minimize (or the negative of the metric you wish to maximize). Here is a pseudo-code example to illustrate this concept:
from sklearn.ensemble import GradientBoostingClassifier from sklearn.model_selection import cross_val_score # Suppose we have some data loaded in X and y X, y = load_your_data() def objective_function(params): # params is a list of hyperparameters learning_rate, n_estimators, criterion = params # Create and train the model with the provided hyperparameters model = GradientBoostingClassifier(learning_rate=learning_rate, n_estimators=n_estimators, criterion=criterion) # Evaluate the model using cross-validation score = cross_val_score(model, X, y, cv=5).mean() # Since we want to maximize the cross-validation score (which is accuracy), # but Bayesian optimization frameworks typically minimize the objective, # we return the negative of the score return -score
In the code example above, the function objective_function is the one we want to minimize. Note we are using 5-fold cross validation in this example, but you could do any type of validation you want, including a simple train-test split.
In Optuna, the search space is defined directly within the objective function, where each hyperparameter is specified using Optuna's domain-specific language (DSL). Hyperparameters are defined individually through methods such as suggest_float for continuous parameters, suggest_int for integer parameters, and suggest_categorical for choosing from a set of categorical options. Each of these methods takes arguments that define the range and characteristics of the hyperparameter, tailored to the specific needs of the optimization task.
Here is an example of defining a search space:
import optuna # Define the hyperparameters search space learning_rate = trial.suggest_float("learning_rate", 0.01, 0.2) n_estimators = trial.suggest_int("n_estimators", 50, 1000) criterion = trial.suggest_categorical("criterion", ["friedman_mse", "mse", "mae"])
In this example, we have three hyperparameters: learning_rate which is a continuous value sampled in a log-uniform way between 0.01 and 0.1, n_estimators which is an integer value between 10 and 100, and criterion which can be "friedman_mse", "mse" or "mae".
The use of prior="log-uniform" for the learning rate means that we initially believe that all scales of learning rate within this range are equally likely to be useful, but we are interested in resolving smaller values with higher precision.
However, it's crucial to understand that this is just a prior belief. As Bayesian optimization proceeds, it utilizes the data collected about the function's performance under different hyperparameters to update this belief. The actual search for hyperparameters, driven by the acquisition function, won't remain uniform but will be more focused on regions that are likely to offer better results according to the updated belief, also known as the posterior.
This approach to exploration and exploitation allows the Bayesian optimization process to efficiently navigate the search space.
The sampler in Optuna refers to the method used to search through the hyperparameter space. The TPESampler performs Bayesian Optimization using Tree-structured Parzen Estimators.
The sampler aims to find the minimum value of the optimization function (often the validation error of the machine learning model) within the given search space, taking into account both the prior distribution of the hyperparameters and the observations made so far to update the posterior belief about the function.
Let's demonstrate Bayesian Optimization using Optuna with an example.
Suppose we are trying to find the minimum of the function 𝑓(𝑥)=(𝑥−2)^2.
Here, 𝑥 is the parameter that we are trying to optimize so that we get the minimum value for 𝑓(𝑥).
This is a simple enough example that we could just use basic derivative calculus to find the minimum of this function: take the derivative of 𝑓(𝑥) with respect to 𝑥, set it equal to zero, and solve for 𝑥. The best value of 𝑥 is 2.0 yielding 𝑓∗(𝑥)=0.
However, let's assume that we don't know the function 𝑓(𝑥) - it's a black box to us. We can only sample from it - meaning, we can only evaluate the function at different values of 𝑥 and observe the output. We want to find the minimum of 𝑓(𝑥) by sampling it at different values of 𝑥, and to do it in as few samples as possible. So, we will use Optuna to solve this optimization problem. Here is the code:
import optuna # Define the function to optimize def objective(trial): # Define the search space for x x = trial.suggest_float('x', -10, 10) return (x - 2) ** 2 # Create a study object that aims to minimize the objective function study = optuna.create_study(direction='minimize') # Optimize the study, specifying the number of optimization calls study.optimize(objective, n_trials=200) # Get the best parameters and the achieved minimum value best_params = study.best_params best_value = study.best_value print("Best value of x: ", best_params['x']) print("Minimum of (x-2)^2: ", best_value)
output:
Best value of x: 2.0006210254762364
Minimum of (x-2)^2: 3.856726421346927e-07
In this example, TPESampler performs Bayesian optimization over the given search space to find the value of 𝑥 that minimizes the function 𝑓(𝑥). We are using 200 trials to find the optimum value of 𝑥.The search results indicate that the best value of 𝑥 is 2.0006, which is very close to the actual optimal value of 2.0. The minimum value of 𝑓(𝑥) is 3.856e-07, which is very close to 0.
This is a toy example to understand the basic concept. In actual machine learning applications, the function to optimize would often be more complex, and the search space would typically contain multiple dimensions representing different hyperparameters. The function to optimize would return the validation error of the machine learning model for a given set of hyperparameters.
1- Define a reasonable hyperparameter space: Hyperparameters can take on any value, but not all values are meaningful. For example, a learning rate might be best within a certain range, and the number of layers in a neural network must be a positive integer. Therefore, defining a reasonable hyperparameter space can help speed up the tuning process and improve results.
2- Start with random or grid search: If you have no idea where to start, random search or grid search can be a good starting point for hyperparameter tuning. These methods can give you a rough idea of what values work best, which can help you narrow down the hyperparameter space for more advanced tuning methods.
3- Use Bayesian Optimization wisely: Bayesian optimization is an advanced technique that can find optimal hyperparameters efficiently, but it also requires careful setup. You need to define the objective function, acquisition function, and surrogate model. Also, Bayesian optimization can take a long time to run, so it may not be the best choice for quick and dirty experiments.
4- Re-evaluate periodically: As you run more experiments and collect more data, it's a good idea to re-run hyperparameter tuning to see if the optimal hyperparameters have changed.
5- Save your results: Always record the results of your hyperparameter tuning, including the hyperparameters tested and the performance of the model. This information can be invaluable for future tuning efforts and for troubleshooting any problems.
6- Ensure a robust evaluation process: The robustness of the evaluation process is vital when tuning hyperparameters. If minor changes in the dataset lead to drastically different loss values, then the evaluation process is not robust and can lead to erroneous conclusions about the optimal hyperparameters. Techniques such as cross-validation can be used to help ensure that the selected hyperparameters perform well on unseen data, providing a more robust and reliable evaluation.
import torch import optuna import torch.nn as nn import torch.optim as optim from torch.utils.data import DataLoader, TensorDataset from sklearn.datasets import load_breast_cancer from sklearn.model_selection import train_test_split from sklearn.preprocessing import StandardScaler from optuna.visualization import plot_optimization_history, plot_param_importances # Load the Breast Cancer dataset data = load_breast_cancer() X, y = data.data, data.target X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42) # Standardize the data scaler = StandardScaler() X_train = scaler.fit_transform(X_train) X_test = scaler.transform(X_test) # Convert to PyTorch tensors X_train = torch.Tensor(X_train) X_test = torch.Tensor(X_test) y_train = torch.Tensor(y_train).long() y_test = torch.Tensor(y_test).long() # Create TensorDatasets and DataLoaders train_dataset = TensorDataset(X_train, y_train) test_dataset = TensorDataset(X_test, y_test) train_loader = DataLoader(train_dataset, batch_size=64, shuffle=True) test_loader = DataLoader(test_dataset, batch_size=64, shuffle=False) # Define the Neural Network architecture class BreastCancerNet(nn.Module): def __init__(self, num_units, num_layers): super(BreastCancerNet, self).__init__() layers = [] input_dim = X_train.shape[1] for _ in range(num_layers): layers.append(nn.Linear(input_dim, num_units)) layers.append(nn.ReLU()) input_dim = num_units # Set input_dim to num_units for the next layer layers.append(nn.Linear(num_units, 2)) # Binary classification self.layers = nn.Sequential(*layers) def forward(self, x): return self.layers(x) # Define the training and evaluation function def train_and_evaluate(model, train_loader, test_loader, optimizer, criterion, device): model.to(device) model.train() for data, target in train_loader: data, target = data.to(device), target.to(device) optimizer.zero_grad() output = model(data) loss = criterion(output, target) loss.backward() optimizer.step() model.eval() correct = 0 with torch.no_grad(): for data, target in test_loader: data, target = data.to(device), target.to(device) output = model(data) pred = output.argmax(dim=1, keepdim=True) correct += pred.eq(target.view_as(pred)).sum().item() accuracy = correct / len(test_loader.dataset) return accuracy # Define the objective function for Optuna def objective(trial): # Suggest values for the hyperparameters num_layers = trial.suggest_int('num_layers', 1, 5) num_units = trial.suggest_int('num_units', 10, 100) lr = trial.suggest_loguniform('lr', 1e-5, 1e-1) model = BreastCancerNet(num_units=num_units, num_layers=num_layers) optimizer = optim.Adam(model.parameters(), lr=lr) criterion = nn.CrossEntropyLoss() # Use a GPU if available device = torch.device("cuda" if torch.cuda.is_available() else "cpu") # Train and evaluate the model accuracy = train_and_evaluate(model, train_loader, test_loader, optimizer, criterion, device) return accuracy # Create the Optuna study and start the optimization study = optuna.create_study(direction='maximize', sampler=optuna.samplers.TPESampler()) study.optimize(objective, n_trials=20) print("Best trial:") trial = study.best_trial print(f" Accuracy: {trial.value:.4f}") print(" Params: ") for key, value in trial.params.items(): print(f" {key}: {value}") # Plot the optimization history fig1 = plot_optimization_history(study) fig1.show() # Plot the parameter importances fig2 = plot_param_importances(study) fig2.show()
output:
[I 2024-08-01 17:52:30,443] Trial 0 finished with value: 0.6228070175438597 and parameters: {'num_layers': 3, 'num_units': 10, 'lr': 0.004363460865583532}. Best is trial 0 with value: 0.6228070175438597.
[I 2024-08-01 17:52:30,451] Trial 1 finished with value: 0.6228070175438597 and parameters: {'num_layers': 3, 'num_units': 77, 'lr': 1.842598326858712e-05}. Best is trial 0 with value: 0.6228070175438597.
[I 2024-08-01 17:52:30,462] Trial 2 finished with value: 0.9385964912280702 and parameters: {'num_layers': 5, 'num_units': 55, 'lr': 0.003609795782359115}. Best is trial 2 with value: 0.9385964912280702.
[I 2024-08-01 17:52:30,468] Trial 3 finished with value: 0.8771929824561403 and parameters: {'num_layers': 1, 'num_units': 47, 'lr': 0.0021827521866422095}. Best is trial 2 with value: 0.9385964912280702.
[I 2024-08-01 17:52:30,474] Trial 4 finished with value: 0.9912280701754386 and parameters: {'num_layers': 2, 'num_units': 11, 'lr': 0.034868374724776115}. Best is trial 4 with value: 0.9912280701754386.
[I 2024-08-01 17:52:30,484] Trial 5 finished with value: 0.9736842105263158 and parameters: {'num_layers': 5, 'num_units': 78, 'lr': 0.039365893118632374}. Best is trial 4 with value: 0.9912280701754386.
[I 2024-08-01 17:52:30,493] Trial 6 finished with value: 0.7719298245614035 and parameters: {'num_layers': 3, 'num_units': 51, 'lr': 4.1074627423742965e-05}. Best is trial 4 with value: 0.9912280701754386.
[I 2024-08-01 17:52:30,501] Trial 7 finished with value: 0.6228070175438597 and parameters: {'num_layers': 5, 'num_units': 20, 'lr': 1.2775137699933669e-05}. Best is trial 4 with value: 0.9912280701754386.
[I 2024-08-01 17:52:30,508] Trial 8 finished with value: 0.8947368421052632 and parameters: {'num_layers': 2, 'num_units': 78, 'lr': 0.03681168690395871}. Best is trial 4 with value: 0.9912280701754386.
[I 2024-08-01 17:52:30,515] Trial 9 finished with value: 0.631578947368421 and parameters: {'num_layers': 4, 'num_units': 42, 'lr': 0.0008712820687871211}. Best is trial 4 with value: 0.9912280701754386.
[I 2024-08-01 17:52:30,526] Trial 10 finished with value: 0.9473684210526315 and parameters: {'num_layers': 1, 'num_units': 26, 'lr': 0.08606752240712784}. Best is trial 4 with value: 0.9912280701754386.
[I 2024-08-01 17:52:30,537] Trial 11 finished with value: 0.9824561403508771 and parameters: {'num_layers': 2, 'num_units': 91, 'lr': 0.017944767247784325}. Best is trial 4 with value: 0.9912280701754386.
[I 2024-08-01 17:52:30,551] Trial 12 finished with value: 0.9824561403508771 and parameters: {'num_layers': 2, 'num_units': 100, 'lr': 0.011206424000488947}. Best is trial 4 with value: 0.9912280701754386.
[I 2024-08-01 17:52:30,561] Trial 13 finished with value: 0.956140350877193 and parameters: {'num_layers': 2, 'num_units': 97, 'lr': 0.00038158687510912465}. Best is trial 4 with value: 0.9912280701754386.
[I 2024-08-01 17:52:30,572] Trial 14 finished with value: 0.9736842105263158 and parameters: {'num_layers': 2, 'num_units': 38, 'lr': 0.015384014691228767}. Best is trial 4 with value: 0.9912280701754386.
[I 2024-08-01 17:52:30,583] Trial 15 finished with value: 0.8859649122807017 and parameters: {'num_layers': 1, 'num_units': 69, 'lr': 0.0002600382900329717}. Best is trial 4 with value: 0.9912280701754386.
[I 2024-08-01 17:52:30,595] Trial 16 finished with value: 0.9298245614035088 and parameters: {'num_layers': 4, 'num_units': 64, 'lr': 0.09003127627687967}. Best is trial 4 with value: 0.9912280701754386.
[I 2024-08-01 17:52:30,607] Trial 17 finished with value: 0.956140350877193 and parameters: {'num_layers': 2, 'num_units': 89, 'lr': 0.012919008952086327}. Best is trial 4 with value: 0.9912280701754386.
[I 2024-08-01 17:52:30,618] Trial 18 finished with value: 0.956140350877193 and parameters: {'num_layers': 3, 'num_units': 31, 'lr': 0.026241644811039317}. Best is trial 4 with value: 0.9912280701754386.
[I 2024-08-01 17:52:30,628] Trial 19 finished with value: 0.9649122807017544 and parameters: {'num_layers': 1, 'num_units': 63, 'lr': 0.008288502687247774}. Best is trial 4 with value: 0.9912280701754386.
Best trial:
Accuracy: 0.9912
Params:
num_layers: 2
num_units: 11
lr: 0.034868374724776115
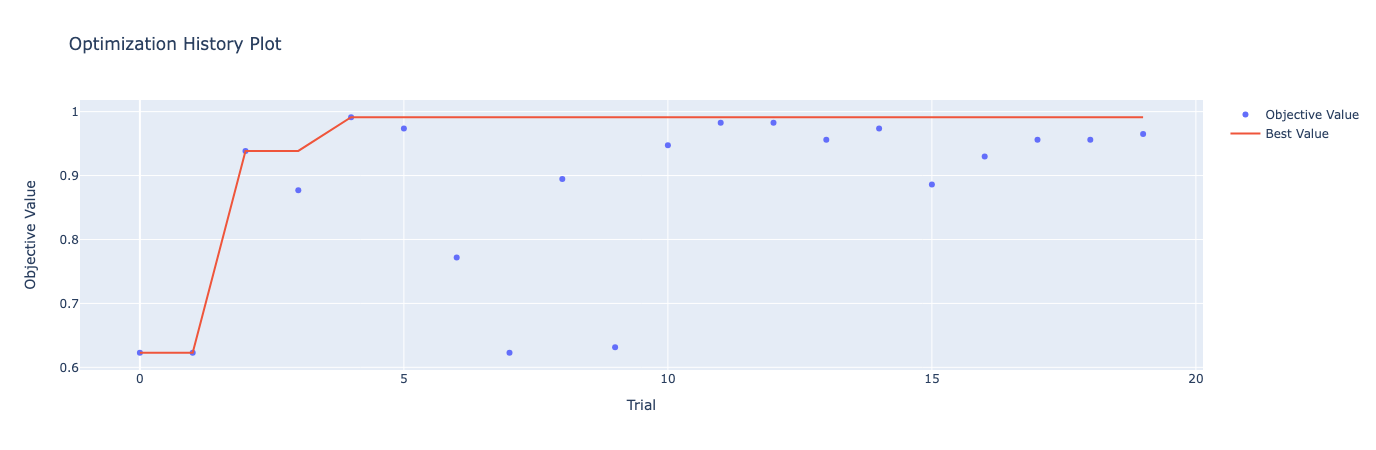
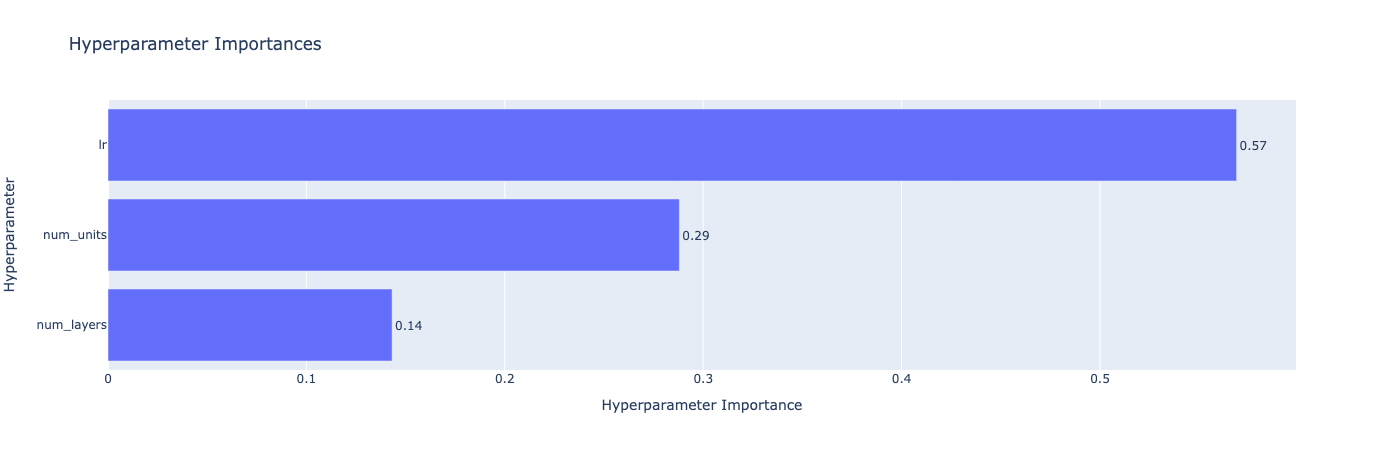
This tutorial guided you through setting up hyperparameter tuning using Bayesian Optimization, defining the objective function, specifying the hyperparameter space, and running the tuning process. Key takeaways included the importance of defining an appropriate hyperparameter space, wisely utilizing Bayesian Optimization, and the necessity of diligent results tracking for future tuning efforts.