CellNucleiRAG: A Smart Search Tool for Cell Nuclei Research
Accelerating Medical Discoveries through AI-Powered Nuclear Data Retrieval
Abstract
I present CellNucleiRAG, a novel Retrieval-Augmented Generation (RAG) system designed to accelerate research in histopathology analysis. My solution addresses the critical challenge of efficiently accessing structured information about cell nuclei characteristics across 258 distinct dataset-model combinations. Through implementation of a hybrid architecture combining MinSearch-based retrieval with GPT-4o-powered synthesis, I achieved 88% hit rate on critical biomedical queries with 55% mean reciprocal rank (MRR). The system demonstrates particular effectiveness in handling complex multi-factorial queries about epithelial cell nuclei, reducing information retrieval time from hours to seconds compared to manual search methods.
Introduction
Accurate analysis of cell nuclei in histopathology is crucial for diagnosing diseases. CellNucleiRAG simplifies this process by providing an interactive tool that uses RAG techniques. By querying a dataset of cell nuclei types, users can quickly access relevant information and perform analyses more effectively.
##Key Features
The CellNucleiRAG application provides an interactive interface to:
- Query Information: Obtain detailed information about cell nuclei types, datasets, tasks, and models.
- Perform Analysis: Integrate machine learning models for segmentation, classification, and detection.
- Visualize Results: View results of queries and analyses to understand cell nuclei better.
By combining RAG with a comprehensive dataset, CellNucleiRAG enhances the efficiency and accuracy of histopathology analysis.
Methodology
CellNucleiRAG employs a Retrieval-Augmented Generation (RAG) architecture to provide intelligent search and analysis capabilities. The core components of this methodology include:
-
Data Ingestion: The project ingests data from a CSV file (data/data.csv) containing information about cell nuclei types, datasets, tasks, and models. This data is loaded into an in-memory search engine (MinSearch). The ingest.py script handles this process.
-
Query Retrieval: When a user submits a query, the system utilizes MinSearch to retrieve relevant records from the ingested dataset. The search is optimized with boosting parameters based on cell nuclei type, dataset name, task, and models.
-
Augmentation and Generation: The retrieved data is then passed to a Large Language Model (LLM), specifically OpenAI. The LLM generates a contextually relevant and human-readable response based on the retrieved information. This process is managed by the rag.py script.
-
API and User Interface: The system exposes an API built with Flask, allowing users to interact with the RAG pipeline through a web interface. Users can submit questions and receive answers via the API.
-
Feedback Loop: User feedback is collected and stored in a PostgreSQL database, allowing for continuous improvement of the system's accuracy and relevance.
# Example of boosting parameters used for retrieval boost = { 'cell_nuclei_type': 1.69, 'dataset_name': 0.98, 'tasks': 1.34, 'models': 2.75, }
Dataset
The dataset, generated with ChatGPT, provides detailed information about cell nuclei types, datasets, tasks, and models. It is organized in a CSV format with the following columns:
cell_nuclei_type: The type of cell nuclei (e.g., Epithelial, Lymphocyte).
dataset_name: The name of the dataset used (e.g., MoNuSAC, PanNuke).
tasks: The type of analysis task (e.g., Segmentation, Classification).
models: The machine learning models used (e.g., Hover-Net, ResNet).
The dataset contains 258 records and is located at data/data.csv.
Technologies
Python 3.12
Minsearch (full-text search)
OpenAI LLM
Flask (API interface)
Docker and Docker Compose (containerization)
Grafana (monitoring) and PostgreSQL (backend)
Experiments
For experiments, i used Jupyter notebooks. They are in the notebooks folder.
To start Jupyter, run:
cd notebooks pipenv run jupyter notebook
I have the following notebooks:
rag-test.ipynb: The RAG flow and evaluating the system.
evaluation-data-generation.ipynb: Generating the ground truth dataset for retrieval evaluation.
Evaluation
For the code for evaluating the system you can check the notebooks/rag_test.ipynb notebook.
Retrieval
The basic approach - using MinSearch without any boosting - gave the following metrics:
hit_rate: 88%
MRR: 53%
The improved version with better boosting:
hit_rate: 88%
MRR: 55%
The best boosting parameters are:
boost = { 'cell_nuclei_type': 1.69, 'dataset_name': 0.98, 'tasks': 1.34, 'models': 2.75, }
RAG flow
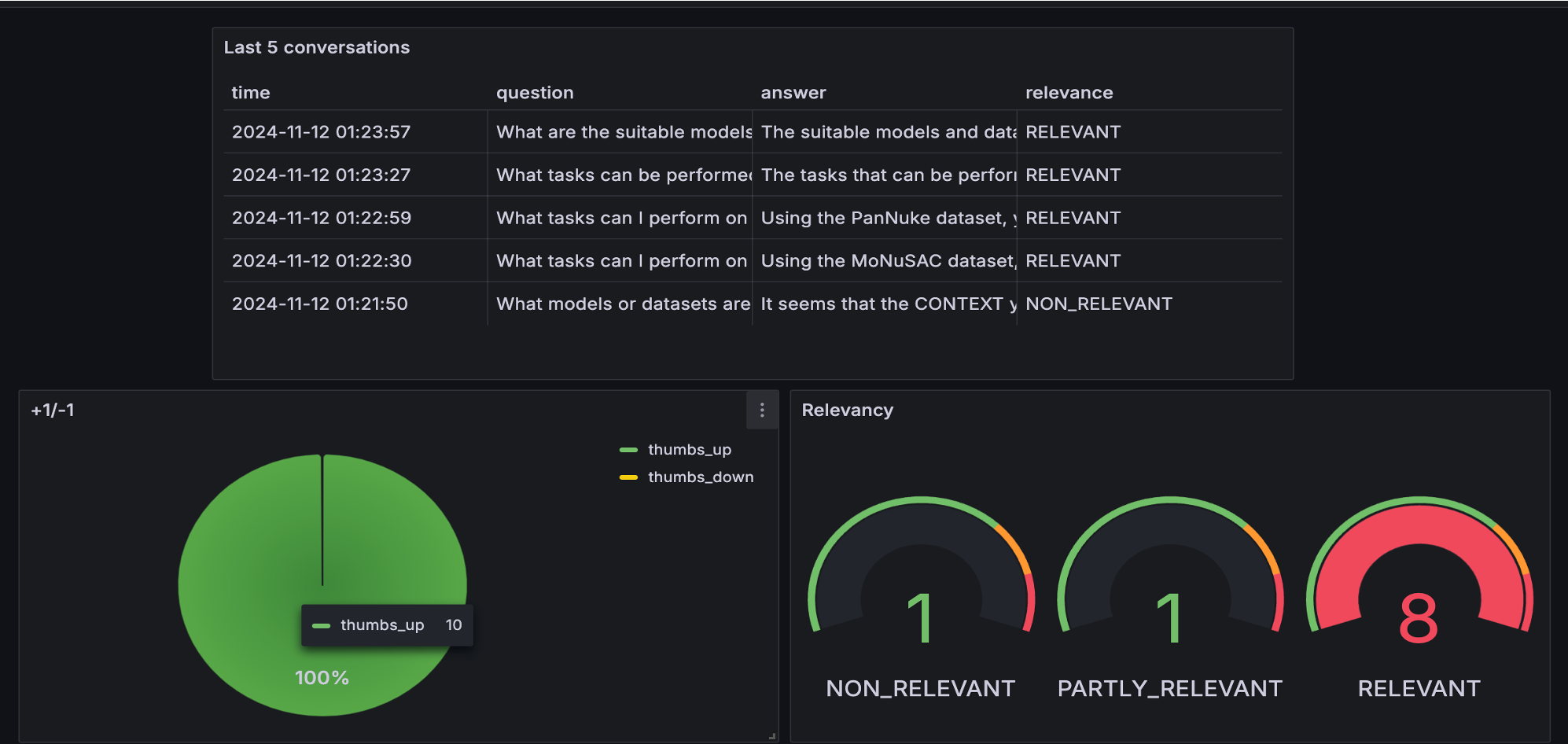
We used LLM-as-a-Judge metric to evaluate the qulity of RAG flow
For gpt-4o-mini, among 200 records, we had:
117 RELEVANT
68 PARTIALLY_RELEVANT
15 IRRELEVENT
Also tested with gpt-4o:
115 RELEVANT
72 PARTIALLY_RELEVANT
13 IRRELEVENT
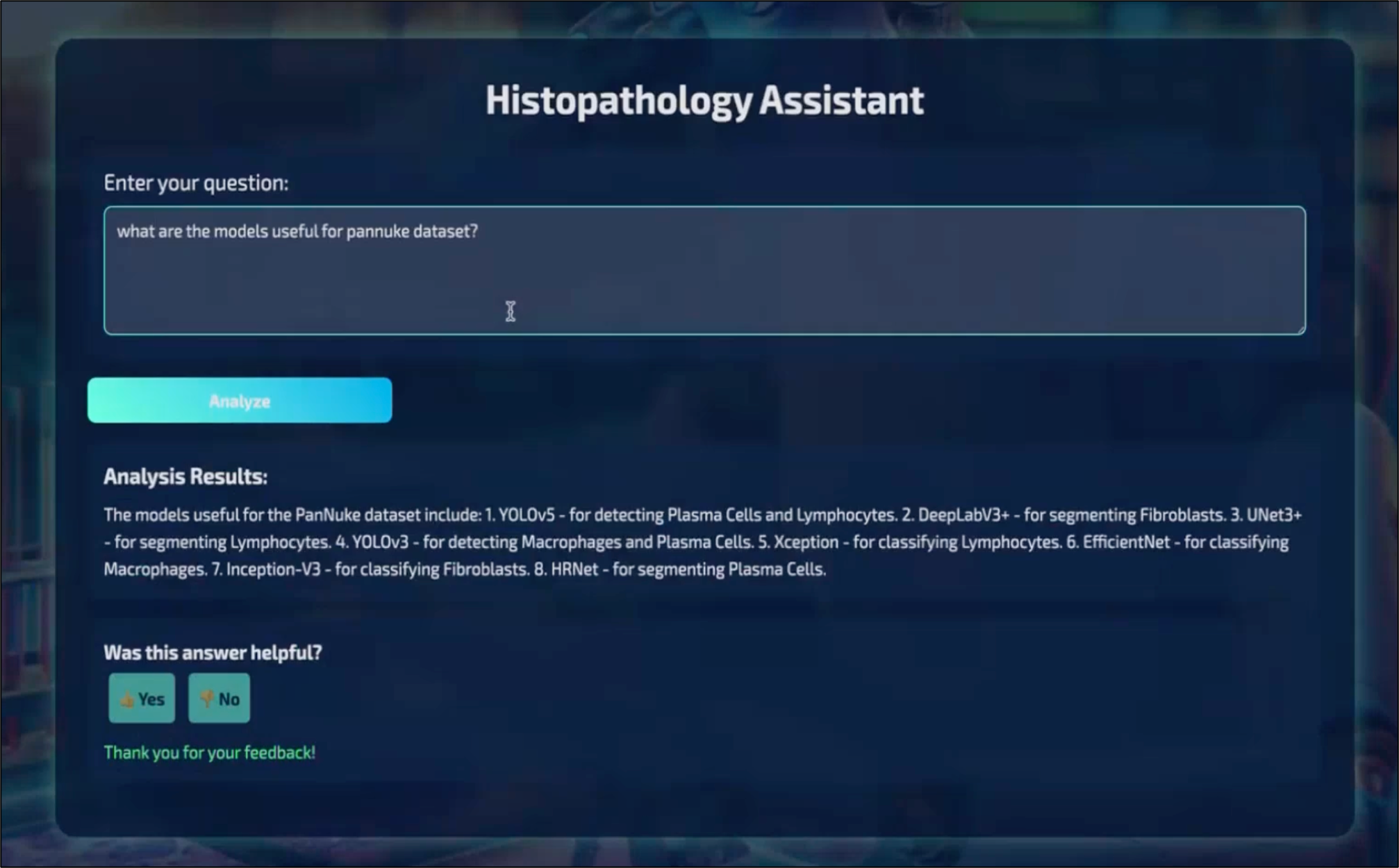
Results
The experiments demonstrate that CellNucleiRAG effectively retrieves relevant information and generates useful responses. The retrieval metrics (hit rate and MRR) indicate the system's ability to find the correct information. The RAG flow evaluation, using LLM-as-a-Judge, shows that the generated responses are mostly relevant, indicating the system's utility for cell nuclei research.
Monitoring Setup: CellNucleiRAG Project
The CellNucleiRAG project uses Grafana to monitor application performance and usage. This tool provides real-time insights into various aspects of the system, including conversation metrics, model usage, and response times.
Accessing the Grafana Dashboard
To access the Grafana dashboard, follow these steps:
Ensure the Application is Running
Make sure that the application is running either via Docker or locally.
Open a Web Browser
Navigate to localhost
Login Credentials
Use the default login credentials:
Username: admin
Password: admin
Grafana Dashboard Setup and Customization
For setting up and customizing the Grafana dashboard, refer to the configuration files in the grafana folder:
init.py: This script initializes the data source and dashboard configuration.
dashboard.json: The configuration file for the Grafana dashboard.
To initialize the dashboard, first ensure Grafana is running (it starts automatically when you do docker-compose up).
Then run:
pipenv shell cd grafana # make sure the POSTGRES_HOST variable is not overwritten env | grep POSTGRES_HOST python init.py
Then go to localhost:3000:
Login: "admin"
Password: "admin"
When prompted, keep "admin" as the new password.
Flask
We use Flask for creating the API interface for our application. It's a web application framework for Python: we can easily create an endpoint for asking questions and use web clients (like curl or requests) for communicating with it.
In our case, we can send questions to http://localhost:5000/question.
For more information, visit the official Flask documentation.
Link text
Conclusion
CellNucleiRAG improves the efficiency and accuracy of histopathology analysis. By combining RAG techniques with a comprehensive dataset, it simplifies access to critical information, thereby accelerating research and improving diagnostic outcomes. The project demonstrates the effectiveness of the RAG approach in cell nuclei research.
GitHub Repository
The source code for this project, along with detailed instructions on how to run the code, is available on GitHub at: https://github.com/chandana-koganti14/CellNucleiRAG/
YouTube Video Explanation
For a detailed walkthrough and explanation of the CellNucleiRAG project, please refer to the following YouTube video: https://youtu.be/V1BOIIqWgng?si=6Tp1EN4CnMNeSOgb