U-Net, Attention U-Net, and Ensemble for Image Segmentation
Project Snapshot
This project explores medical image segmentation using U-Net and Attention U-Net models, focusing on blood vessel structures in kidney images. By combining predictions from both models into an ensemble, the aim is to enhance segmentation accuracy and provide robust results.
Key Findings and Results
Performance Metrics:
-
U-Net:
- Best Epoch: 11
- Training Loss: 0.0958
- Validation Loss: 0.1106
- Training Dice Coefficient: 0.917
- Validation Dice Coefficient: 0.905
-
Attention U-Net:
- Best Epoch: 19
- Training Loss: 0.2705
- Validation Loss: 0.2824
- Training Dice Coefficient: 0.778
- Validation Dice Coefficient: 0.771
-
Ensemble Model:
- Training Loss: 0.1832
- Validation Loss: 0.1965
- Training Dice Coefficient: 0.848
- Validation Dice Coefficient: 0.838
Observations:
- The U-Net outperforms the Attention U-Net overall, achieving higher Dice Coefficients and better generalization on validation data.
- The original U-Net was best for his expirment. Ensembling lowered the overall accuracy.
- The loss function demonstrates the extreme data imbalance in the dataset, as seen in the class counts:
- Class 0 (Background): 26,059,006
- Class 1 (Blood Vessels): 155,394
- Class 1 to Class 0 Ratio: 0.00596
- Higher losses in initial epochs indicate challenges in learning underrepresented vessel structures.
Dataset
- Name: Blood Vessel Segmentation Dataset
- Source: Kaggle Competition
- Structure:
- Training Images
- Validation Images
- Testing Images
Visual Results
The following visualization provides a comparison of the segmentation results. The images show the original medical image, the true mask (ground truth), and the predicted mask from the segmentation model. These demonstrate the effectiveness of the U-Net and Attention U-Net models in capturing vessel structures:
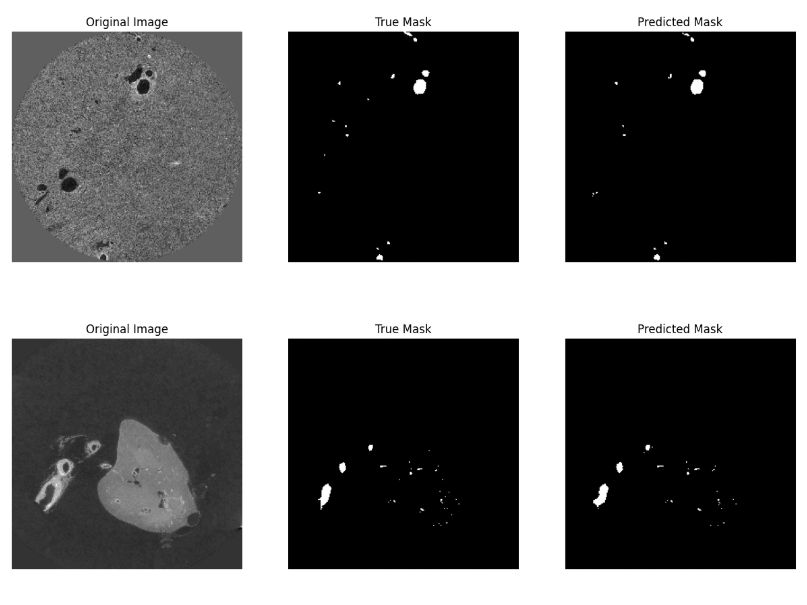
Methodology
Models:
-
U-Net:
- Backbone: ResNet50 (pre-trained on ImageNet).
- Focuses on hierarchical feature extraction.
-
Attention U-Net:
- Adds attention gates to suppress irrelevant regions in images, enhancing vessel boundary detection.
-
Ensemble:
- Combines predictions from both models through weighted averaging to improve robustness.
Loss Function:
- Binary Cross-Entropy + Dice Coefficient:
- Ensures both pixel-wise accuracy and segmentation quality.
- Demonstrates the impact of data imbalance, highlighting challenges in learning underrepresented structures.
How to Reproduce
In Kaggle:
- Open the notebook in the Kaggle environment:
Sennet U-Net, Attention U-Net, and Ensemble - Enable GPU acceleration.
- Click "Run All" to execute the notebook.
Locally:
- Clone the repository.
- Download the dataset from Kaggle:
Blood Vessel Segmentation Dataset - Install required dependencies:
pip install tensorflow keras spacy nltk opencv-python-headless matplotlib - Modify paths for local execution.
- Run the notebook.
Tools and Technologies
- Frameworks: TensorFlow, Keras
- Languages: Python
- Dependencies: OpenCV, Matplotlib
- Environment: Kaggle (with GPU acceleration)
Insights and Challenges
Insights:
- The U-Net demonstrates superior performance over the Attention U-Net, achieving higher Dice Coefficients.
- Ensemble models provide a balanced trade-off between precision and recall.
Challenges:
- Training with limited GPU memory required optimization in data loading and batch processing.
- Extreme data imbalance, with a background-to-vessel ratio of 0.00596, posed significant challenges in model training.
Next Steps
- Explore the use of other segmentation models to ensemble with the original U-Net.
- Incorporate additional datasets for generalization across different medical imaging modalities.
- Deploy the model for real-time segmentation tasks.
For further details, refer to the notebook and associated documentation in the repository.