Introduction to Random Survival Forests (RSF)

Random Survival Forests (RSF) represent an advanced statistical method designed for analyzing survival data, where the primary challenge is dealing with right-censored outcomes. Such data are prevalent in medical and biological studies where the event of interest (e.g., death, relapse) might not occur within the study period for all subjects. Developed from Breiman's Random Forests, RSF adapts and extends the ensemble tree methodology specifically for survival data, enhancing predictive accuracy and interpretability in scenarios plagued by censoring and complex interactions among variables.
Motivation for RSF
Traditional survival analysis techniques, like the Cox proportional hazards model, make stringent assumptions about the hazard functions (e.g., proportional hazards) and often struggle with high-dimensional data, complex interaction effects, and nonlinear relationships. These methods also require explicit model specification, which can be cumbersome and error-prone, particularly when dealing with multifaceted datasets.
In contrast, RSF is a non-parametric method that does not require assumptions about the form of the survival function. It automatically detects interactions and nonlinearities without needing predefined model structures. This flexibility makes RSF a robust tool for survival analysis, providing insights that are often missed by more traditional approaches.
Core Components of RSF
RSF constructs a multitude of decision trees based on bootstrapped samples of the data. Each tree contributes to an ensemble prediction that improves robustness and accuracy. The method involves several key steps and components:
-
Bootstrap Sampling: Each tree in the forest is built from a bootstrap sample, i.e., a randomly selected subset of the data with replacement, allowing some observations to appear multiple times and others not at all.
-
Tree Growing: Unlike standard decision trees that may use criteria like Gini impurity or entropy, survival trees in an RSF use split criteria based on survival differences, effectively handling censored data.
-
Splitting Criteria: The survival split is based on maximizing the differences in survival outcomes between groups formed at each node. This involves calculating a split statistic that measures how well the split separates individuals with different survival prospects.
-
Random Feature Selection: At each split in the tree, a random subset of features is considered. This randomness injects diversity into the models, reducing the variance of the ensemble prediction.
-
Ensemble Predictions: After many trees are grown, predictions for individual observations are made by averaging results across the forest. This ensemble approach helps in reducing the variance and avoiding overfitting.
-
Cumulative Hazard Function: RSF models estimate the cumulative hazard function for each individual, providing a direct way to interpret the survival function.
Mathematical Foundation of RSF
The mathematical underpinning of RSF involves concepts from survival analysis and decision tree algorithms:
-
Survival Trees: Each tree is built by optimizing a survival-related objective function, typically involving the hazard function or survival times directly.
-
Nelson-Aalen Estimator: In terminal nodes of each tree, the cumulative hazard is estimated using the Nelson-Aalen estimator, a non-parametric statistic that accumulates hazard over time.
-
Conservation of Events: A key theoretical property of RSF is the conservation of events principle, ensuring that the total number of events predicted by the forest equals the number observed in the data.
This introduction sets the stage for a deeper exploration into the technical aspects and applications of RSF, illustrating how this powerful method provides substantial advancements over traditional models in handling complex survival data.
Random Survival Forests Algorithm
-
Draw Bootstrap Samples:
- B bootstrap samples are drawn from the original dataset.
- Each bootstrap sample excludes on average 37% of the data. This excluded portion is referred to as out-of-bag (OOB) data.
-
Grow a Survival Tree for Each Bootstrap Sample:
- At each node within a tree, p candidate variables are randomly selected.
- The node is then split using the candidate variable that maximizes the survival difference between the resulting daughter nodes.
-
Tree Growth Constraints:
- Each tree is grown to its full size with the constraint that each terminal node must contain no fewer than d0 unique death events.
-
Calculate Cumulative Hazard Function (CHF):
- A CHF is calculated for each tree.
- These CHFs are then averaged across all trees to obtain the ensemble CHF.
-
Prediction Error Calculation:
- Using the OOB data, the prediction error for the ensemble CHF is calculated.
This algorithmic structure allows the RSF method to effectively deal with the complexities of right-censored survival data, utilizing the ensemble of trees to improve prediction accuracy and stability.
Binary Survival Trees
Binary survival trees, a core component of the Random Survival Forests (RSF) algorithm, are specialized decision trees tailored for survival analysis. Their structure and growth process are analogous to Classification and Regression Trees (CART), but they are specifically optimized for handling right-censored survival data. Here's a brief overview of how binary survival trees function:
Growth Process
- Initialization: The process begins at the root node, which contains all the dataset's observations.
- Node Splitting: This root node is then recursively split into two daughter nodes (left and right), based on a predetermined survival criterion.
- Recursive Splitting: Each of these daughter nodes is similarly split, giving rise to further left and right nodes. This recursive splitting continues for each subsequent node down the tree.
Splitting Criteria
- Maximizing Survival Differences: The key to splitting a node is to find a partition that maximizes the survival difference between the resulting daughter nodes. This involves evaluating all potential variables (denoted as
) and their possible split points (denoted as ). - Selection of Split Points: The optimal split point (
) and variable ( ) are those that create the most significant survival difference between the two new groups. This criterion helps in effectively separating dissimilar cases.
Goal of Tree Growth
- Homogeneity: As the tree grows and branches further, the aim is to increase homogeneity within each node. That is, each terminal node eventually comprises cases that are similar in terms of survival characteristics, making the predictions more accurate and tailored to specific groups within the data.
Binary survival trees leverage the natural variability in survival data to form predictions that are robust and capable of capturing complex interactions and non-linear relationships inherent in such datasets. The final tree structure provides a nuanced and insightful model of survival probability distributions tailored to various covariate patterns observed in the data.
Terminal Node Prediction
In Random Survival Forests (RSF), terminal node prediction plays a crucial role in estimating the survival function for individual cases within the dataset. By grouping similar cases into terminal nodes and calculating the cumulative hazard function (CHF) for each node, RSF provides a detailed and nuanced approach to survival analysis. Here's a detailed look at how terminal node prediction works in RSF:
Terminal Node Formation
- Criterion for Terminal Nodes: Each node in a survival tree must contain a minimum number of unique death events, denoted as
. This criterion ensures that statistical conclusions drawn from each terminal node are reliable and supported by sufficient data. - Saturation Point: A survival tree reaches saturation when no new splits can be formed that meet the criteria, making further data division unfeasible. At this point, the most extreme nodes are termed as terminal nodes.
Data in Terminal Nodes
- Survival and Censoring Data: Each terminal node, denoted by
, contains survival times and censoring status for each individual in the node. These are crucial for calculating survival statistics. - If
, the individual is right-censored, meaning the event of interest (e.g., death) has not occurred by the end of the study period. - If
, it indicates that the event of interest has occurred.
- If
Calculating the Cumulative Hazard Function (CHF)
- Distinct Event Times: Within each terminal node, all distinct event times are noted, and these are used to compute the cumulative hazard for the node.
- Nelson-Aalen Estimator: The CHF estimate for each terminal node is computed using the Nelson-Aalen estimator. This is given by the formula:
Here,represents the number of deaths at time , and is the number of individuals at risk at that time.
CHF for Individual Cases
- Covariate Influence: Each case
in the dataset is associated with a set of covariates . The CHF for an individual case that falls into terminal node is the CHF calculated for that node:
- Homogeneity Within Nodes: All cases within a terminal node
share the same CHF estimate, implying that cases grouped together are statistically similar concerning their survival characteristics.
Terminal node prediction in RSF is a methodically rigorous process that focuses on capturing and utilizing the survival characteristics of cases grouped into the most granular segments (terminal nodes) of the tree. By calculating a CHF using the Nelson-Aalen estimator, RSF provides a robust method to estimate the survival function for groups of similar cases, allowing for nuanced and precise survival analysis. This methodology ensures that the survival predictions are not only accurate but also reflect the complex interactions of covariates in a right-censored survival context.
The Bootstrap and Out-Of-Bag (OOB) Ensemble Cumulative Hazard Function (CHF) in Random Survival Forests
The process of calculating the ensemble Cumulative Hazard Function (CHF) in Random Survival Forests (RSF) involves two distinct types of estimates: the Out-Of-Bag (OOB) ensemble CHF and the bootstrap ensemble CHF. These calculations leverage the strengths of the bootstrap sampling and the ensemble nature of RSF to provide robust survival predictions.
Out-Of-Bag (OOB) Ensemble CHF
The OOB ensemble CHF is particularly crucial as it provides an unbiased performance estimate of the RSF model, since it is calculated using data not seen during the training of individual trees.
-
Definition and Calculation:
- Each tree in the RSF is grown from a unique bootstrap sample of the original dataset. Any data point not included in this bootstrap sample for a particular tree is considered OOB for that tree.
- For a data point
, indicate whether it is OOB for tree using an indicator function , where if is OOB for ; otherwise, . - The CHF for a tree
grown from the bootstrap sample, evaluated at a set of covariates , is denoted by . - The OOB ensemble CHF for data point
is then calculated as a weighted average of the CHFs from those trees where is OOB, weighted by the indicator function:
This formula essentially averages the CHF predictions across all bootstrap samples where the data point
is OOB, providing a CHF estimate based on trees that did not use in their training process.
Bootstrap Ensemble CHF
Unlike the OOB estimate, the bootstrap ensemble CHF uses all trees in the forest to predict the CHF for each data point, providing a comprehensive average that includes all bootstrap variations.
-
Calculation:
- The bootstrap ensemble CHF for a data point
is the simple average of the CHFs from all trees in the forest:
This method utilizes all survival trees, not just those where the data point
is OOB. It integrates the predictions from every single tree in the ensemble, providing a robust measure that considers all the potential splits and variable importance assessments captured across the entire forest. - The bootstrap ensemble CHF for a data point
Significance and Usage
- OOB Ensemble CHF: Offers an unbiased estimate of the model's prediction error and is often used for model validation and tuning parameters.
- Bootstrap Ensemble CHF: Provides a comprehensive prediction using the entire forest, often used for the final model predictions after the optimal parameters are determined.
The dual approach of using both OOB and bootstrap ensemble CHFs allows RSF to leverage the benefits of bootstrap aggregating (bagging) to reduce variance, enhance prediction accuracy, and avoid overfitting, making it highly effective for survival analysis in various complex datasets.
Understanding the C-index Calculation in Random Survival Forests
The C-index, or concordance index, is a crucial statistical tool used to evaluate the predictive accuracy of survival models, particularly in the presence of censored data. It is analogous to the area under the Receiver Operating Characteristic (ROC) curve used in other types of predictive modeling. Here's an in-depth look at how the C-index is calculated in the context of Random Survival Forests (RSF):
Purpose of the C-index
The C-index estimates the probability that, in a randomly selected pair of cases, the case that experiences the event of interest (e.g., death) first had a worse predicted outcome (higher risk score). This measure is particularly useful in survival analysis as it inherently accounts for right-censoring and does not rely on a fixed follow-up time, making it a flexible and robust measure of model performance.
Steps to Calculate the C-index
-
Form All Possible Pairs of Cases:
- The first step involves creating all possible pairs from the dataset. Each pair consists of two different cases.
-
Omit Censored Pairs:
- Pairs where the survival time of both cases is censored are omitted because it’s unclear when the events would have occurred.
- Additionally, pairs where the survival times are equal (i.e.,
) are also omitted unless one of the events is a death. This ensures that comparisons are made between distinguishable outcomes.
-
Counting Concordant Pairs:
- For pairs where
, a count of 1 is added if the case with the shorter survival time has a worse predicted outcome (higher risk of the event). This means the prediction aligns with the actual outcome. - If the predicted outcomes are tied but the survival times differ, a count of 0.5 is added, indicating partial concordance.
- For pairs where
and both cases are deaths, a count of 1 is added if the predicted outcomes are tied, reflecting perfect concordance in predicting the event at the same time. - If
but not both are deaths, a count of 1 is added if the case that died has a worse predicted outcome; otherwise, a count of 0.5 is added.
- For pairs where
-
Calculation of the C-index:
- The final C-index,
, is calculated by dividing the total concordance score by the number of permissible pairs (those pairs included in the analysis after the above omissions). Mathematically, it is expressed as:
- This ratio provides a value between 0 and 1, where 1 indicates perfect predictive accuracy, and 0.5 suggests no better accuracy than random chance.
- The final C-index,
Significance of the C-index
The C-index is particularly valued in medical statistics and survival analysis because it provides a direct and interpretable measure of a model's predictive ability concerning the timing of events. Its calculation for RSF models reflects not only the ability to rank individuals by risk but also how well the model handles censored data, a common challenge in clinical trials and other longitudinal studies.
By integrating the handling of censored data directly into the metric, the C-index provides a comprehensive and realistic assessment of the model's predictive performance in real-world scenarios where not all outcomes are observed.
Results
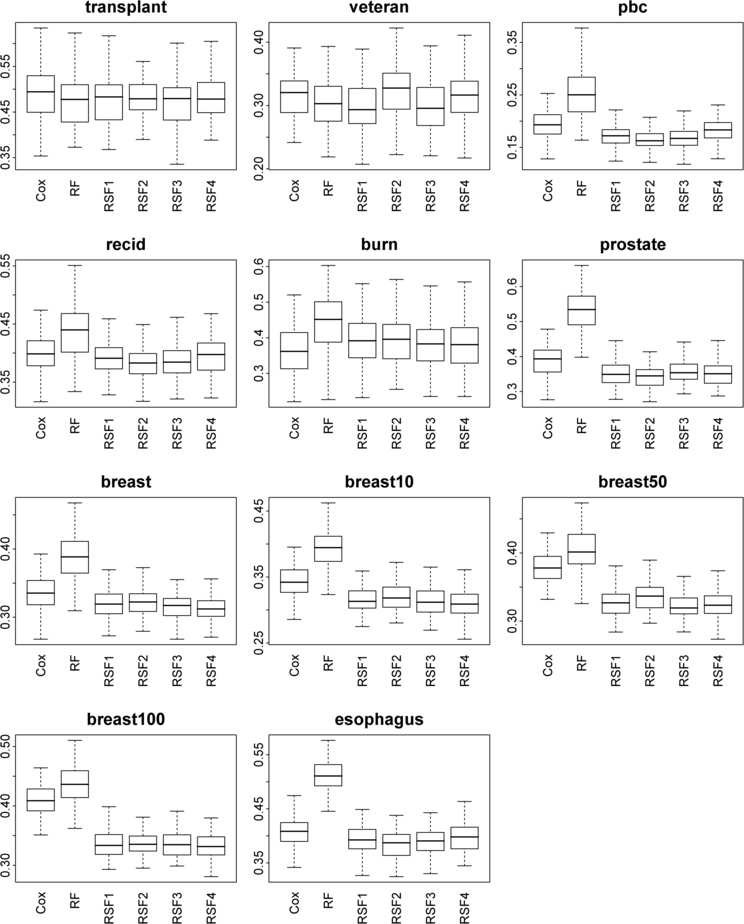
Analysis of Boxplots for Estimated Prediction Error in Random Survival Forests
The provided boxplots represent the estimated prediction error across various datasets, calculated using the C-index, a measure of concordance that reflects how well a model can discriminate between different outcomes in survival analysis. These errors are estimated from 100 independent bootstrap replicates, and the results are shown for different methods including Cox regression, Random Forest (RF) specifically for censored data, and several variations of Random Survival Forests (RSF).
Datasets Analyzed
The analysis includes several datasets, each pertinent to different health and survival scenarios:
- Transplant, Veteran, PBC: These are related to medical studies involving patients with specific conditions.
- Recid, Burn: Involve datasets from criminal recidivism studies and burn patients.
- Prostate: Involves survival analysis on prostate cancer patients.
- Breast, Breast10, Breast50, Breast100: Refer to breast cancer datasets with varying numbers of noise variables added to test the stability of the model against irrelevant features.
- Esophagus: Likely related to survival after esophageal cancer.
Methods Compared
- Cox: Cox regression, a traditional model used in survival analysis.
- RF: Random Forest adapted for censored survival data.
- RSF1 through RSF4: Different configurations of Random Survival Forests:
- RSF1: Utilizes log-rank splitting.
- RSF2: Applies a conservation-of-events principle.
- RSF3: Employs a log-rank score method.
- RSF4: Uses a random log-rank splitting technique.
Observations from the Boxplots
- Variability and Central Tendency: Each boxplot shows the interquartile range, median (horizontal line), and mean (dots) of the prediction errors. The whiskers extend to show the range of the data, providing insight into the variability of each method's performance across the bootstrap samples.
- Comparison Across Methods:
- The Cox model generally shows higher prediction errors compared to RSF methods, suggesting that RSF may handle censored data and complex interactions more effectively.
- RSF variants typically show lower median errors and less variability, indicating more robust and consistent performance. This is particularly evident in datasets with added noise (Breast10, Breast50, Breast100), where RSF's ability to manage irrelevant features shines.
- The RSF models, especially RSF2 (conservation-of-events) and RSF3 (log-rank score), often outperform the standard RF model, highlighting the advantages of RSF's specialized survival analysis techniques.
Conclusion of the Blog on Random Survival Forests (RSF)
In this paper, wRandom Survival Forests (RSF), an innovative extension of Breiman's renowned forest methodology, tailored specifically for the analysis of right-censored survival data. RSF harnesses the power of ensemble learning through the construction of multiple survival trees, each grown from independently drawn bootstrap samples. By randomly selecting subsets of variables at each node and employing a survival-specific splitting criterion based on survival time and censoring information, RSF meticulously adapts to the nuances of survival data.
A key strength of RSF lies in its terminal node analysis, where each node's cumulative hazard function (CHF) is estimated using the Nelson-Aalen estimator, providing a refined measure of the risk at each node. The ensemble CHF, obtained by averaging these estimates across all trees, offers a robust prediction model. Additionally, the use of out-of-bag (OOB) data allows for nearly unbiased estimates of prediction error, making RSF not only powerful but also reliable in its assessments.
The innovative approaches embedded within RSF, including a novel algorithm for handling missing data, enable it to provide almost unbiased error estimates even in scenarios plagued with substantial amounts of incomplete data. This adaptability is crucial for both training and testing phases, enhancing the model's applicability and accuracy across diverse datasets.
Empirical evaluations underscore the superiority of RSF over traditional methods. Across a broad array of real and simulated datasets, RSF has demonstrated consistent superiority or comparable performance against existing methods. Its ability to discern complex interrelationships among variables is particularly notable. For instance, in a case study examining coronary artery disease, RSF was instrumental in elucidating intricate associations among renal function, body mass index, and long-term survival—relationships that have often been obscured or oversimplified in previous studies.
The Variable Importance Measure (VIMP), another innovative feature of RSF, further exemplifies its capability to identify and emphasize significant predictors without the need for extensive manual tuning typical of more conventional models. This facilitates a more automated and insightful exploration of data, revealing critical insights that might otherwise remain hidden.
In conclusion, Random Survival Forests represent a significant advancement in the field of survival analysis, offering a sophisticated yet user-friendly tool that extends the analytical capabilities of researchers and data scientists. By integrating robust statistical techniques with the flexibility of machine learning, RSF stands out as a premier method for tackling the complexities of survival data, providing clear, actionable insights that are vital for scientific advancement and practical application.
References
- Ishwaran, H., Kogalur, U. B., Blackstone, E. H., & Lauer, M. S. (2008). Random survival forests. The annals of applied statistics, 2(3), 841-860.
- Ishwaran, H., & Kogalur, U. B. (2010). Random survival forests for R. R news, 7(2), 25-31.