The primary objective of this project is to explore the application of deep learning in the healthcare sector, specifically through the use of transfer learning with pretrained models such as ResNet18 for the classification of X-ray images. The goal is to develop a model capable of accurately distinguishing between normal and pneumonia-affected images.
Utilizing a ResNet18 pretrained model and a dataset from Kaggle, the model exhibited strong training performance but achieved a modest validation accuracy of 65%, likely due to the limited dataset of 20 data points used for validation. Despite this, the model demonstrated excellent performance on the test set with an accuracy of 95%.
Additionally, the project includes the development and deployment of a user-friendly application on Stream lit, enabling users to upload X-ray images and receive predictions, thereby showcasing the practical utility of the model.
The “Deep Learning for Pneumonia Detection Using X-ray Imaging” project aims to demonstrate the critical role of Artificial Intelligence (AI) in healthcare, specifically in detecting diseases using historical data to train models. These models can significantly aid medical professionals in saving lives and developing strategies to reduce infections such as pneumonia. While the classification approach showcased in this project is focused on healthcare, it is versatile and can be applied in various other sectors including finance, sports, and education.
To overcome resource limitations, such as the lack of GPU-enabled computers, transfer learning with pretrained models like ResNet18 was utilized. The training was conducted on Google Colab, which offers free but limited GPU services. Leveraging pretrained models, trained under optimal conditions, ensures high accuracy—a crucial requirement for health-related outcomes.
This project aims to leverage deep learning techniques for the classification of pneumonia from chest X-ray images. By utilizing advanced convolutional neural networks (CNNs), the study seeks to enhance diagnostic accuracy and support medical professionals in detecting pneumonia efficiently.
The methodology involves data preprocessing, model training using CNN architectures, and performance evaluation on a labeled dataset of X-ray images. Key steps include data augmentation, hyperparameter tuning, and cross-validation to ensure robustness. Below are some of the processes:
import os import numpy as np import pandas as pd from PIL import Image from tqdm.auto import tqdm from matplotlib import pyplot as plt from sklearn.model_selection import train_test_split from sklearn.metrics import ConfusionMatrixDisplay, confusion_matrix import torch from torchinfo import summary import torch.nn.functional as F from torch.optim.lr_scheduler import StepLR import torch.nn as nn from torch.optim import Adam from torch.utils.data import Dataset, DataLoader, random_split from torchvision import transforms, datasets from torchvision import models from torchvision.models import resnet18
# define data directories train_path = ("/content/drive/MyDrive/chest_xray/train") test_path = ("/content/drive/MyDrive/chest_xray/test") val_path = ("/content/drive/MyDrive/chest_xray/val")
# get classes from the training data classes = os.listdir(train_path) print('There are', len(classes), 'classes') print(classes)
# converting images to RGB class ConvertToRGB(): def __call__(self, img): if img.mode != 'RGB': img = img.convert("RGB") return img # defining transforms transform = transforms.Compose( [ ConvertToRGB(), transforms.Resize((224, 224)), transforms.ToTensor(), ] )
# creating data sets using ImageFolder train_dataset = datasets.ImageFolder(root = train_path, transform = transform) val_dataset = datasets.ImageFolder(root = val_path, transform = transform) test_dataset = datasets.ImageFolder(root = test_path, transform = transform) # to be used for testing print(train_dataset) print(val_dataset)
# introducing a pretrained model model = models.resnet18(weights= models.ResNet18_Weights.DEFAULT) print(model)
# training function def train_model(model, train_loader, val_loader, loss_fn, optimizer, num_epochs, scheduler, early_stopping, checkpointing, device = 'cpu' ): for epoch in range(num_epochs): print(f"Epoch {epoch+1}/{num_epochs}") print('-' * 10) # Iterate over training and validation phases for phase in ['train', 'val']: if phase == 'train': model.train() # Set model to training mode data_loader = train_loader else: model.eval() # Set model to evaluation mode data_loader = val_loader running_loss = 0.0 running_corrects = 0 # Iterate over data for inputs, labels in data_loader: inputs, labels = inputs.to(device), labels.to(device) optimizer.zero_grad() # Forward pass with torch.set_grad_enabled(phase == 'train'): outputs = model(inputs) _, preds = torch.max(outputs, 1) loss = loss_fn(outputs, labels) # Backward pass + optimization only in training phase if phase == 'train': loss.backward() optimizer.step() # Statistics running_loss += loss.item() * inputs.size(0) running_corrects += torch.sum(preds == labels.data) epoch_loss = running_loss / len(data_loader.dataset) epoch_acc = running_corrects.double() / len(data_loader.dataset) print(f'{phase}_Loss: {epoch_loss:.4f} {phase}_Acc: {epoch_acc:.4f}')
#load the model and evaluate the test data model.load_state_dict(torch.load("pneumonia_model.pth", weights_only = True)) model.to(device) model.eval() y_true, y_pred =[], [] with torch.no_grad(): for images, labels in test_loader: images, labels = images.to(device), labels.to(device) outputs = model(images) _, preds = torch.max(outputs,1) y_true.extend(labels.cpu().numpy()) y_pred.extend(preds.cpu().numpy()) accuracy = accuracy_score(y_true, y_pred) print(f'The model accuracy on unseen data is', accuracy)
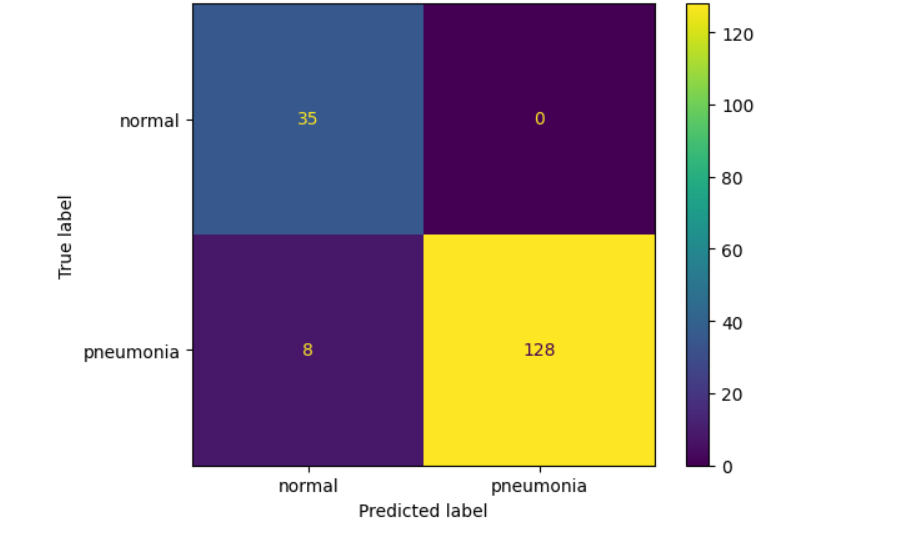
The model accuracy on unseen data is 0.9532163742690059
disp = ConfusionMatrixDisplay(matrix_test, display_labels = ['normal', 'pneumonia']) disp.plot()
def test_prediction(model, dataset,i): image = dataset[i][0] image_batch= image.unsqueeze(0) model = model.to(device) with torch.no_grad(): image_batch = image_batch.to(device) output = model(image_batch) _, predicted_class = output.max(1) # mapping the predcition to the class class_names = ['normal', 'pneumonia'] # Map predictions to class names predicted_labels = [class_names[idx] for idx in predicted_class.cpu().numpy()] # Convert to list of class names #Print results for idx, label in enumerate(predicted_labels): print(f"The predicted label for image {i + 1} is '{label}'.") # displaying the image image = np.transpose(image, (1, 2, 0)) image = np.clip(image, 0, 1) plt.imshow(image) plt.axis('off') predicted_label = predicted_labels[0] plt.text(10, 20 , f'Predicted: {predicted_label}', fontsize = 15, color = "red", bbox=dict(facecolor= 'white', alpha=0.7)) plt.show() predicted_labels =class_names[predicted_class.item()] print('The predicted label is ', predicted_labels)
Pnuemonia-Xray-ImageClassification
The findings indicate that deep learning models, particularly CNNs, can effectively analyze X-ray images to detect pneumonia. Challenges such as dataset bias and model interpretability are discussed, along with potential improvements for future research.
This study demonstrates the potential of deep learning in medical imaging, particularly for pneumonia detection. By integrating AI tools into healthcare, diagnostic efficiency and accuracy can be significantly improved, paving the way for advanced medical solutions.
This report documents the development and deployment of a Streamlit application for pneumonia detection using deep learning models. The project utilizes ResNet-18, a convolutional neural network (CNN), to classify X-ray images as either "Pneumonia" or "Normal." The application aims to provide a user-friendly interface for medical professionals to quickly and accurately assess chest X-rays.
The application integrates a pre-trained ResNet-18 model, fine-tuned for binary classification. Key stages include:
ResNet-18 is loaded with pretrained weights (ResNet18_Weights.DEFAULT) for feature extraction.
The fully connected layer is customized for binary classification with softmax activation.
Input images are converted to RGB (if necessary).
Images are resized to 224x224 pixels and normalized using standard ImageNet mean and standard deviation values.
Uploaded X-ray images are processed through the neural network to output probabilities.
The model determines the class label (Normal or Pneumonia) based on the highest probability.
Visualization:
The original image is displayed alongside the prediction result.
A heatmap overlay is generated for better interpretability.
The application demonstrates reliable performance on test images. It accurately distinguishes between "Normal" and "Pneumonia" cases when provided with valid X-ray images. Results are displayed directly in the Streamlit interface, making it accessible for non-technical users. See the image below:
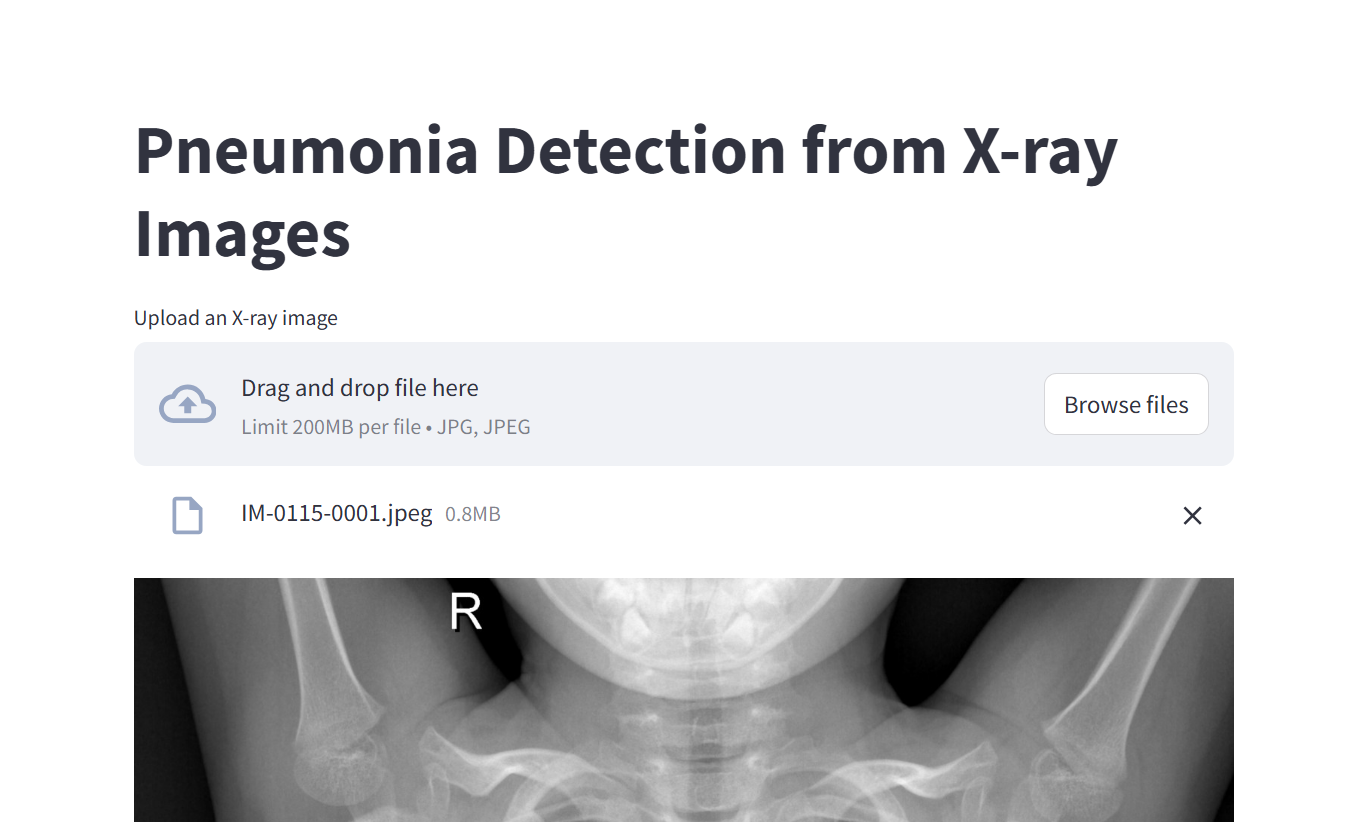
The app interface
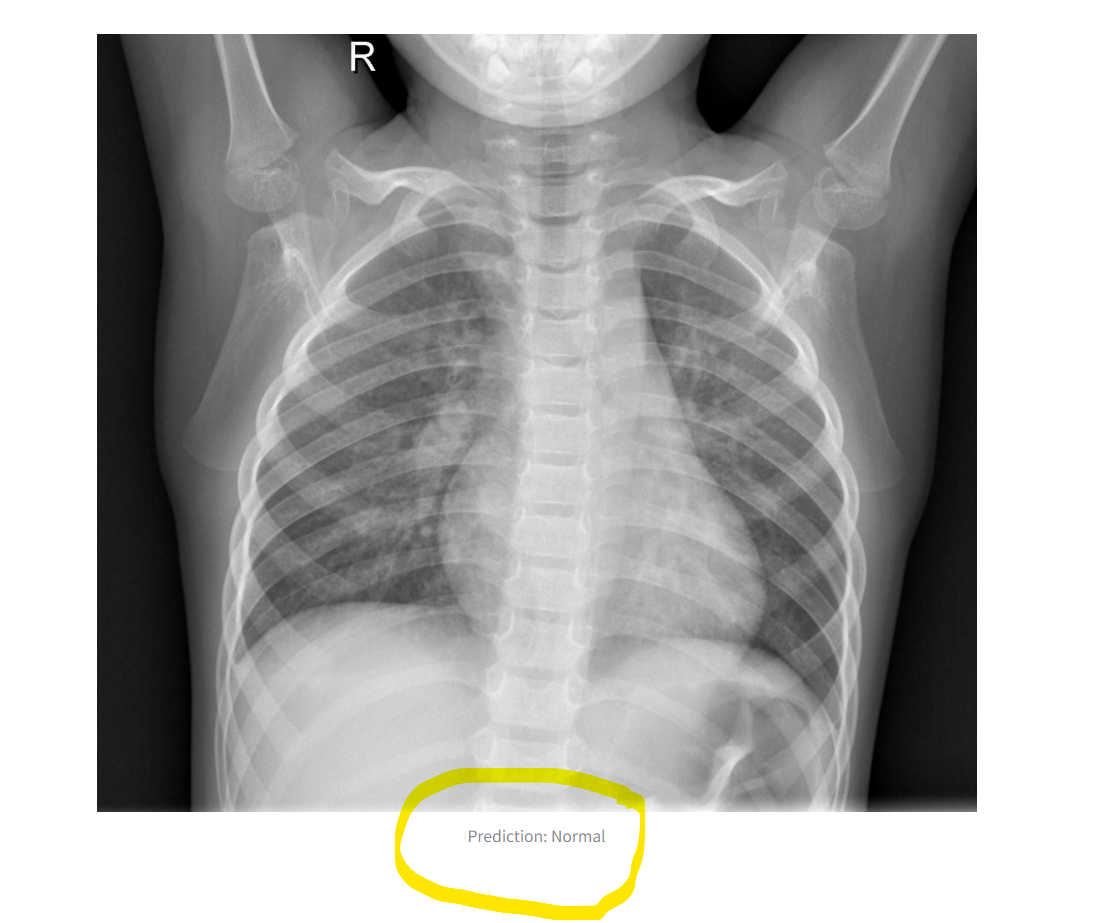
The output image and label
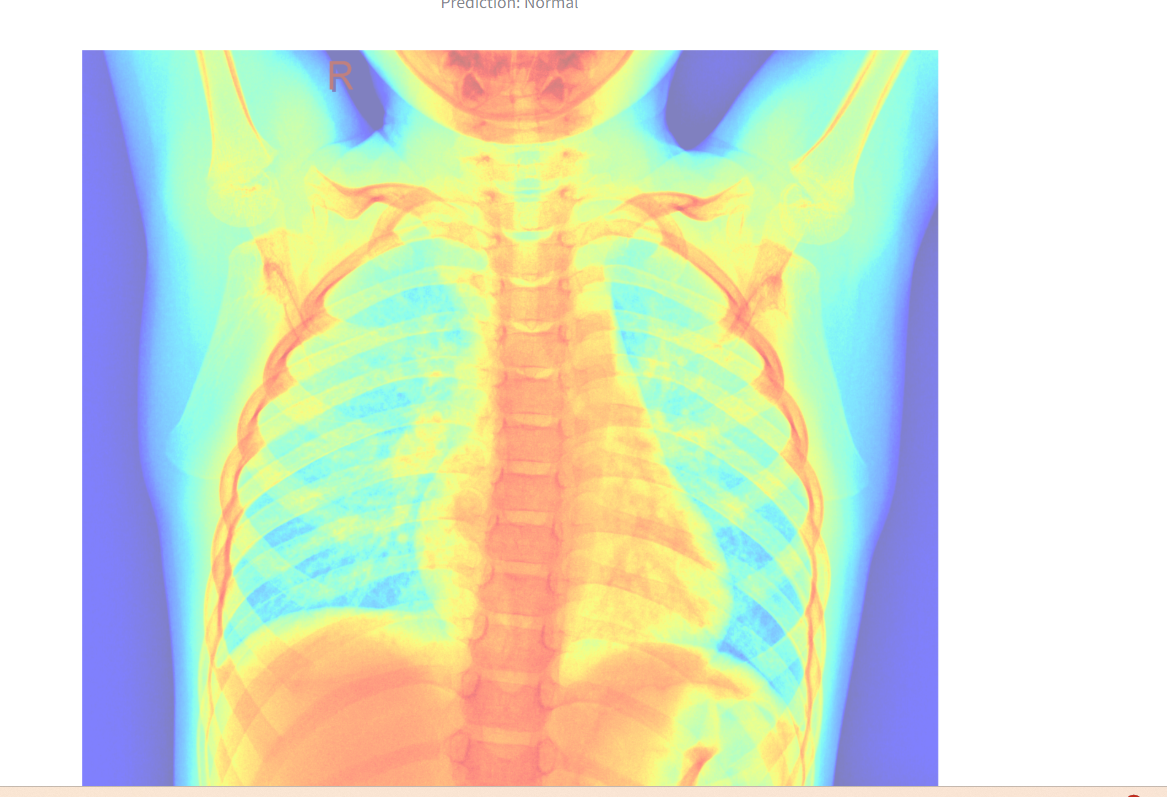
The heatmap overlay
Ease of Use: The Streamlit interface simplifies interaction, allowing users to upload images and receive instant predictions.
Performance: Leveraging a fine-tuned ResNet-18 model ensures robust feature extraction and classification accuracy.
Visualization: The app enhances interpretability with a heatmap overlay.
Generalization: The model’s performance is reliant on the quality and diversity of the training dataset.
Device Limitation: Computations were performed on a CPU, which might affect processing time for large datasets.
Expanding the dataset to improve generalization across diverse X-ray imaging conditions.
Incorporating explainability tools such as Grad-CAM for enhanced interpretability.
This Streamlit application effectively applies deep learning to classify pneumonia from X-ray images. By offering a straightforward interface and reliable predictions, it demonstrates the practical potential of AI in healthcare diagnostics.