This project focuses on developing a Convolutional Neural Network (CNN) to classify brain tumor MRI scans into four categories: Healthy Brain, Glioma Tumor, Meningioma Tumor, and Pituitary Tumor. The primary goal is to facilitate early diagnosis by leveraging deep learning technology to improve the accuracy of MRI image classification. Furthermore,
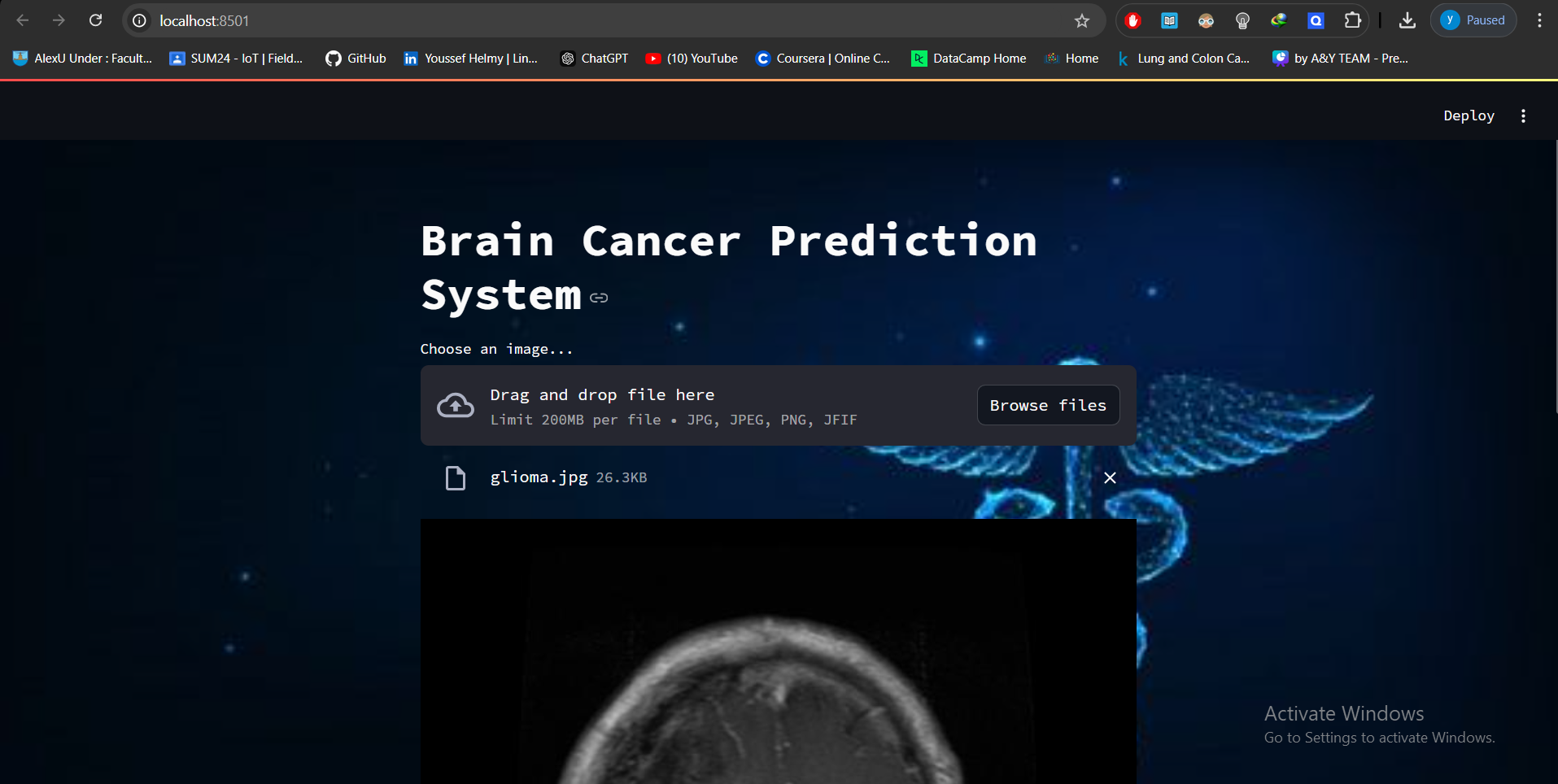
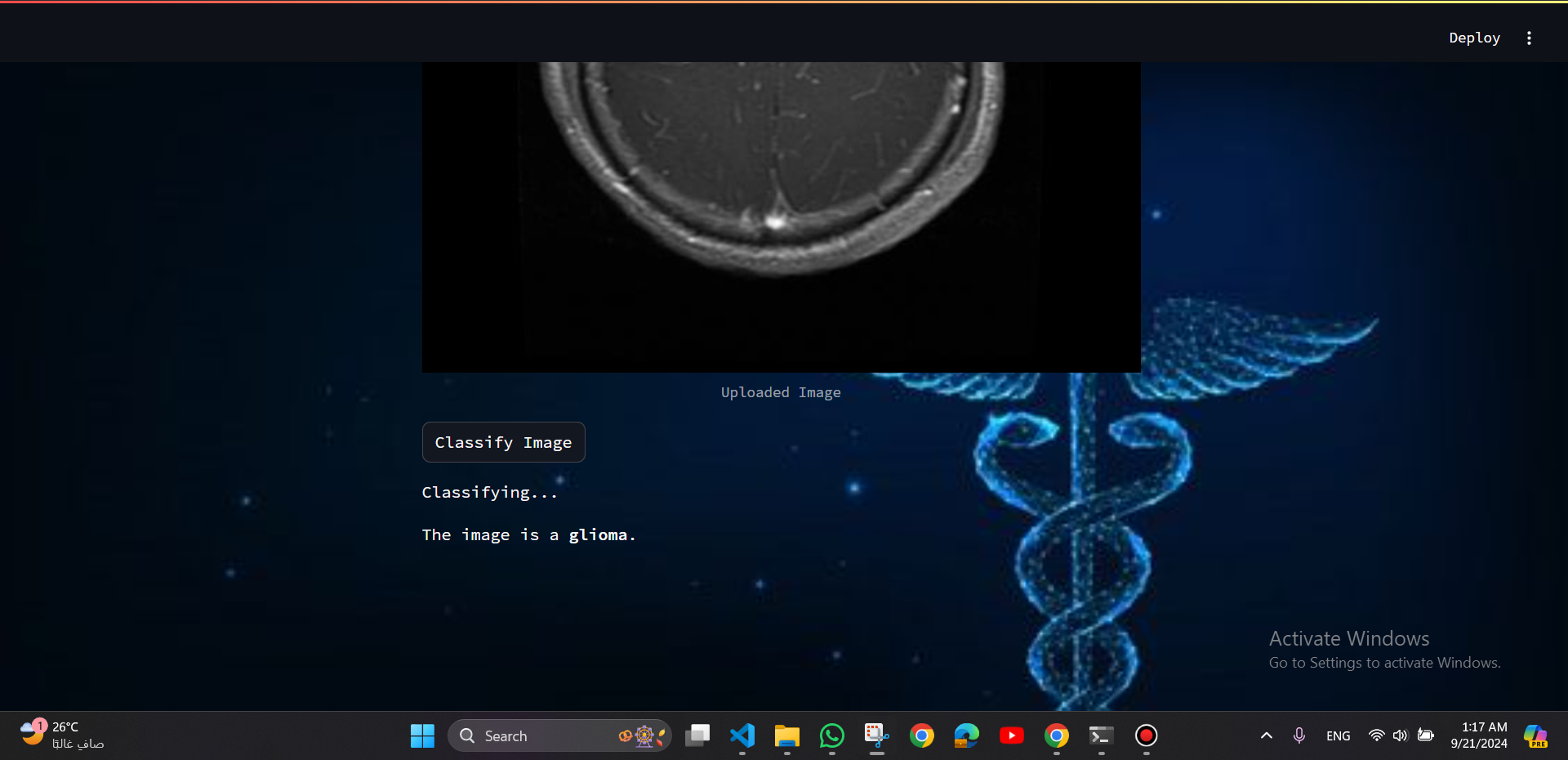
a Streamlit web app was developed to provide real-time classification, offering a user-friendly interface for doctors to enhance the diagnostic process.
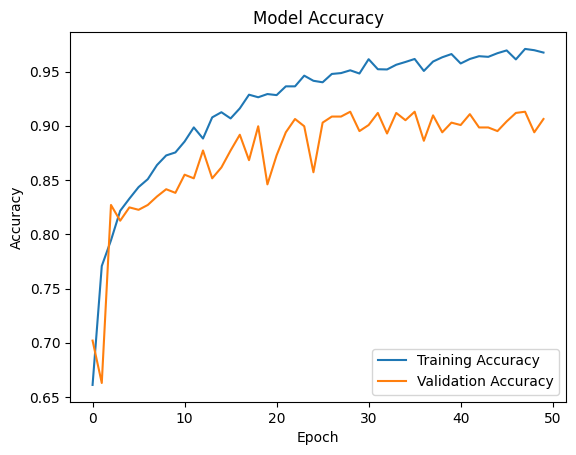
The CNN model achieved an accuracy of 97% on the training dataset and 93% on the testing dataset, demonstrating its reliability and effectiveness in distinguishing between different types of brain tumors.
The dataset used for this project was sourced from Kaggle, which includes MRI scans categorized into four classes:
The dataset consists of a diverse set of images with varying resolutions and orientations, requiring preprocessing for uniformity.
The preprocessing steps involved the following:
A Convolutional Neural Network (CNN) architecture was developed to classify the MRI images. The model comprises the following layers:
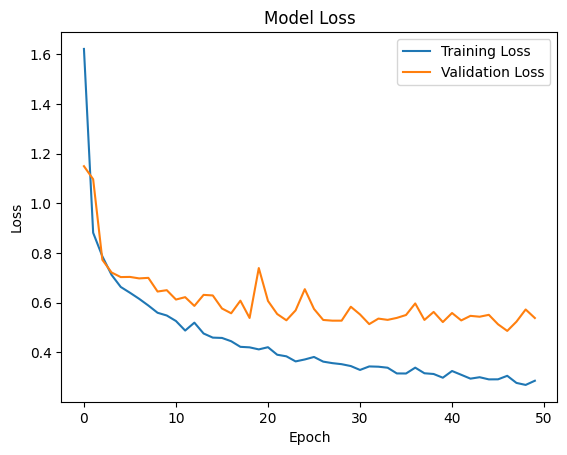
The model was trained using the Adam optimizer and Sparse Categorical Crossentropy as the loss function. The training involved 50 epochs with a batch size of 32, and a validation split of 15% of the training data was used to monitor the model's performance during training.
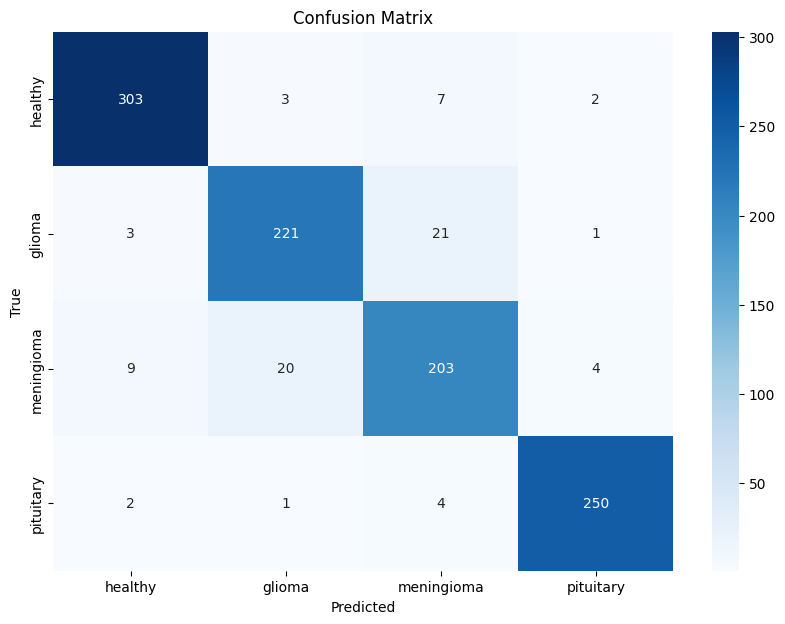
The trained CNN was evaluated on the test set, where it achieved an accuracy of 93%. Additionally, a confusion matrix and classification report were generated to analyze the model's performance across each class.
The CNN model achieved 97% accuracy on the training data and 93% accuracy on the test data, proving the effectiveness of the architecture.
The confusion matrix and classification report revealed that the model performed well across all classes, with minor misclassifications between certain tumor types.
To enhance the accessibility and utility of the model, it was integrated into a Streamlit web application. The web app provides an intuitive interface for users to upload MRI images and receive real-time predictions regarding the presence of brain tumors.
is a promising open-source Python library, which enables developers to build attractive user interfaces in no time. Streamlit is the easiest way especially for people with no front-end knowledge to put their code into a web application: No front-end (html, js, css) experience or knowledge is required.
This project successfully developed and deployed a deep learning-based brain tumor classification system using CNN and integrated it into a Streamlit web application for real-time classification. The model's high accuracy highlights its potential utility in aiding medical professionals in diagnosing brain tumors more efficiently.
import matplotlib.pyplot as plt import numpy as np import cv2 import os import PIL import tensorflow as tf import pickle import pathlib from sklearn.model_selection import train_test_split from tensorflow import keras from tensorflow.keras import layers from tensorflow.keras.regularizers import l2 from tensorflow.keras.models import Sequential
from google.colab import files uploaded= files.upload() for fn in uploaded.keys(): print('User uploaded file "{name}" with length {length} bytes'.format( name=fn, length=len(uploaded[fn]))) !mkdir -p ~/.kaggle && mv kaggle.json ~/.kaggle/ && chmod 600 ~/.kaggle/kaggle.json
!kaggle datasets download -d rm1000/brain-tumor-mri-scans
!unzip brain-tumor-mri-scans.zip -d brain_tumor
data_dir = pathlib.Path('/content/brain_tumor')
list(data_dir.glob('*/*.jpg'))[:5]
image_count = len(list(data_dir.glob('*/*.jpg'))) print(image_count)
glioma = list(data_dir.glob('glioma/*')) PIL.Image.open(str(glioma[0]))
healthy= list(data_dir.glob('healthy/*')) PIL.Image.open(str(healthy[0]))
meningioma= list(data_dir.glob('meningioma/*')) PIL.Image.open(str(meningioma[0]))
pituitary= list(data_dir.glob('pituitary/*')) PIL.Image.open(str(pituitary[0]))
# Count the number of images in each class glioma_count = len(list(data_dir.glob('glioma/*.jpg'))) healthy_count = len(list(data_dir.glob('healthy/*.jpg'))) meningioma_count = len(list(data_dir.glob('meningioma/*.jpg'))) pituitary_count = len(list(data_dir.glob('pituitary/*.jpg'))) # Create a pie chart labels = ['Brain Glioma', 'Healthy Brain', 'Meningioma Brain','Pituitary Brain'] sizes = [glioma_count, healthy_count,meningioma_count ,pituitary_count ] colors = ['red', 'green', 'yellow','pink'] explode = (0.1, 0.1, 0.1,0) # explode the 1st slice plt.pie(sizes, explode=explode, labels=labels, colors=colors, autopct='%1.1f%%', shadow=True, startangle=140) plt.axis('equal') # Equal aspect ratio ensures that pie is drawn as a circle. plt.title('Distribution of Brain Cancer MRI Images') plt.show()
brain_images_dict = { 'healthy': list(data_dir.glob('healthy/*')), 'glioma': list(data_dir.glob('glioma/*')), 'meningioma': list(data_dir.glob('meningioma/*')), 'pituitary': list(data_dir.glob('pituitary/*')), }
brain_labels_dict = { 'healthy': 0, 'glioma': 1, 'meningioma': 2, 'pituitary':3, }
X, y = [], [] for diagnosis, images in brain_images_dict.items(): for image in images: img = cv2.imread(str(image)) resized_img = cv2.resize(img,(80,80)) X.append(resized_img) y.append(brain_labels_dict[diagnosis])
X = np.array(X) y = np.array(y)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.15 , random_state=42)
X_train_scaled = X_train / 255.0 X_test_scaled = X_test / 255.0
# CNN Architecture with Regularization and Dropout num_classes = 4 model1 = Sequential([ layers.Conv2D(32, 3, padding='same', activation='relu', kernel_regularizer=l2(0.01)), layers.MaxPooling2D(), layers.Conv2D(64, 3, padding='same', activation='relu', kernel_regularizer=l2(0.01)), layers.MaxPooling2D(), layers.Conv2D(128, 3, padding='same', activation='relu', kernel_regularizer=l2(0.01)), layers.MaxPooling2D(), layers.Flatten(), layers.Dense(256, activation='relu', kernel_regularizer=l2(0.01)), layers.Dense(num_classes, activation='softmax') ]) model1.compile(optimizer='adam', loss=tf.keras.losses.SparseCategoricalCrossentropy(), metrics=['accuracy']) # Train the model history = model1.fit(X_train_scaled, y_train, validation_split=0.15, batch_size=32, epochs=50)
model1.evaluate(X_test_scaled,y_test)
predictions = model1.predict(X_test_scaled)
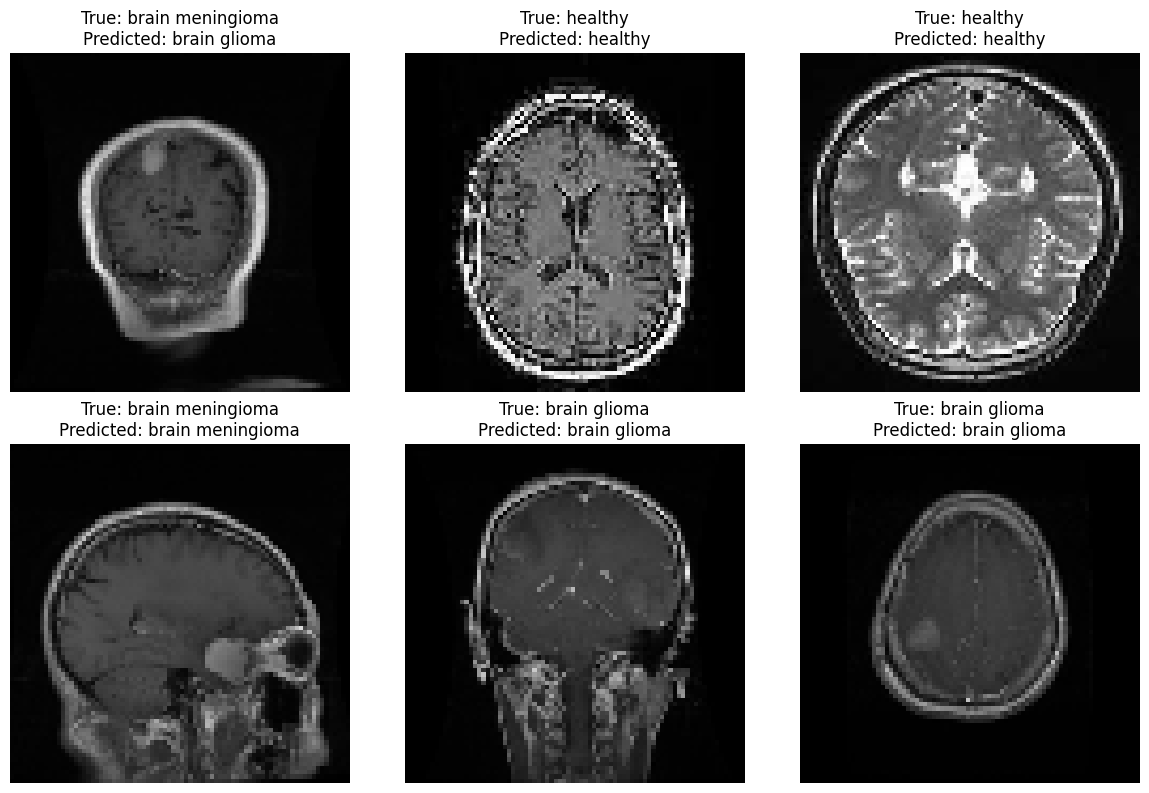
label_to_class = { 0: 'healthy', 1: 'brain glioma', 2: 'brain meningioma', 3: 'brain pituitary', } # Select a random subset of images to display num_images_to_display = 6 random_indices = np.random.choice(len(X_test), num_images_to_display, replace=False) # Create a figure and subplots fig, axes = plt.subplots(2, 3, figsize=(12, 8)) axes = axes.flatten() for i, index in enumerate(random_indices): image = X_test[index] true_label = y_test[index] predicted_label = np.argmax(predictions[index]) axes[i].imshow(image) axes[i].set_title(f"True: {label_to_class[true_label]}\nPredicted: {label_to_class[predicted_label]}") axes[i].axis('off') plt.tight_layout() plt.show()
plt.plot(history.history['accuracy'], label='Training Accuracy') plt.plot(history.history['val_accuracy'], label='Validation Accuracy') plt.title('Model Accuracy') plt.xlabel('Epoch') plt.ylabel('Accuracy') plt.legend(loc='lower right') plt.show()
plt.plot(history.history['loss'], label='Training Loss') plt.plot(history.history['val_loss'], label='Validation Loss') plt.title('Model Loss') plt.xlabel('Epoch') plt.ylabel('Loss') plt.legend(loc='upper right') plt.show()
from sklearn.metrics import classification_report y_pred = np.argmax(predictions, axis=1) print(classification_report(y_test, y_pred, target_names=['healthy', 'brain glioma', 'brain meningioma', 'brain pituitary']))
import matplotlib.pyplot as plt import numpy as np import seaborn as sns from sklearn.metrics import confusion_matrix # Generate predictions for the test set y_pred = np.argmax(model1.predict(X_test_scaled), axis=-1) # Compute confusion matrix cm = confusion_matrix(y_test, y_pred) # Create a heatmap for the confusion matrix plt.figure(figsize=(10, 7)) sns.heatmap(cm, annot=True, fmt='d', cmap='Blues', xticklabels=['healthy', 'glioma', 'meningioma','pituitary'], yticklabels=['healthy', 'glioma', 'meningioma','pituitary']) plt.xlabel('Predicted') plt.ylabel('True') plt.title('Confusion Matrix') plt.show()
model1.save('Brain_Cancer_model.keras')